We characterized N,N-dimethyl formamide (GIF), (PDB), as well as the radicals formed by abstraction of the formyl hydrogen (GIF), (PDB), or one of the methyl hydrogens (GIF), (PDB). We also characterized the dimethyl amino radical (GIF), (PDB), which is formed by breaking of the central CN bond. The calculated energies of these species, together with tabulated G2 energies for H and HCO [5} allow us to calculate the bond dissociation energies (BDE), at 0 K, of various bonds in DMF.
| kcal/mole | |
| HC(O)N(CH3)CH2-H | 75.0 |
| H-C(O)N(CH3)2 | 89.1 |
| HC(O)-N(CH3)2 | 75.3 |
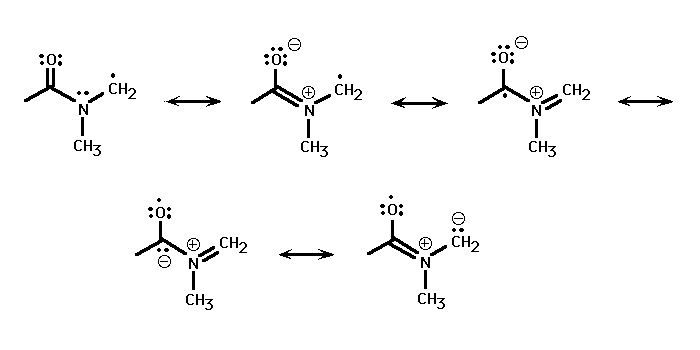
The formyl C-H bond strength is almost unchanged from that in formaldehyde, which has a 0 K bond dissociation energy of 86.6 kcal/mole. [7] In contrast, the methyl C-H bond is considerably weaker than that in dimethyl amine, which has a 298 K bond dissociation energy of 86.5 kcal/mole. [8] The weakening of the methyl C-H bond strength is a result of the HC(O)N(CH3)(CH2) radical's unexpected stability. The radical is found to have a near-planar OCNC backbone; (GIF), (PDB). the electron can then be delocalized throughout the radical backbone.
