
Model 1. Navigation Dashboard for a VRML Model, deriving from the CosmoWorld viewer.
To illustrate the difference between a modelling and a markup language, consider how an atom might be described. HTML itself has no well defined mechanism for defining an atom, and one has to use a markup language such as CML5 (Chemical Markup Language, which is an implementation of XML, itself an evolutionary successor to HTML) to define an atom and properties such as atomic number, its connectivity to other atoms, the number of electrons associated with it and so forth. Using VRML, one would define the same atom as an spherical object, with model properties such as colour, radius, 3D co-ordinates, lighting and motion attributes, and if necessary associate this object with scriptable actions such as collision avoidance with other objects (computed if necessary using e.g. molecular mechanics force fields). VRML is therefore complementary to a markup language since it defines a quite different set of primitive and importantly three-dimensional objects which can be used to express complex chemical models in a visual manner. Copyright implications for models are also different from illustrations, in part because models are specified using data provided by their author, and only created in a specific viewable style by the actions of the reader and the software they are using.
One page-derived metaphor does carry over; the author of a model can insert the equivalent of section headings (called viewpoints) to emphasise what they believe are important characteristics of their model, and in the process also provide the equivalent of keywords ("meta-data") to assist in any indexing of their model. A rather less desirable characteristic of VRML models is that even minor syntactic errors in their specification (the equivalent of spelling mistakes or errors in the markup of an HTML document) will probably render the entire model unviewable, a contrast with component-based HTML documents which fail more gracefully in failing to properly display only the component with the error. This intolerance of errors is probably one reason why the application of VRML is still relatively low.

Creating VRML worlds is more complex than authoring an HTML document, and invariably requires the acquisition of suitable software. A good generic package is CosmoWorlds6, whilst for chemical applications, programs such as WebLab Viewer Pro7 or EyeChem8 are available. Several "filters" for converting standard chemical coordinate formats such as PDB also exist.9
 |
VRML has been applied to many areas of chemistry9. The first VRML models were published by our group in December 1994 (Model 2) resulting from our experiments10 in what we called "molecular collaboratories" using the newly introduced UK high speed national computer network. These models retained the familiar wireframe, ball and stick, spacefill or ribbon representations of molecules, but enabled the rich navigation features of VRML (Model 1), and the ability to define hyperlinks from individual regions of the molecule to other Internet resources, in much the same way as HTML allows such hyperlinks in text-based documents. |
| Model 2. An early VRML model showing the 3CRO protein/DNA complex. |
| Another great advantage of VRML is that it can be readily extended to displaying more complex chemicals such as ionic lattices, large biopolymers, carbohydrates, peptides, liquid crystals etc. These often require different symbolic representations to illustrate the key features. Some excellent examples of this type of application emerged from Brickmann's11 group in Darmstadt from 1995 onwards. Their work shows how more complex models (Model 3)11 can reveal the 3D structure of e.g. the p53 tumor suppressor protein complexed with a DNA helix (blue); the peptide backbone of the protein is rendered as a ribbon, with the computed electrostatic potentials at the interface being projected onto the respective Van der Waals surfaces of the molecules. The red groups are the "mutation hotspots". |
 |
| Model 3. A VRML model illustrating the key features of the p53 tumor suppressor protein (taken from Y. Cho, S. Gorina, and N. Pavletich, Science, 1994, 265, 346). |
 |
Hewat in Grenoble12 showed early on how a VRML representation (Model 4) of the structure of the high-temperature superconductor Y2Ba4Cu7O15, composed of alternating Y1Ba2Cu3 and Y1Ba2Cu4 units could be constructed dynamically from a set of options and parameters provided by the user via a Web-based form. |
| Model 4. VRML representation of a high-temperature superconductor (taken from A. W. Hewat, P. Fischer, E. Kaldis, J. Karpinski, S. Rusiecki and E. Jilek, Physica C, 1990, 167, 579 ). |
 |
Bragg once said that "the important thing in science is not so much to obtain new facts, as to obtain new ways of thinking about them". Knowledge discovery in databases (KDD), or, more popularly, "data mining", has generated a great deal of interest in recent years as vast quantities of information have begun to accumulate in databases. The objective is to be able to trawl these databases in novel ways so as to discover unusual relations and correlations between and within the data. We have applied VRML13 to the problem of representing 3D information "mined" from e.g. the Cambridge Crystallographic Database14 to help identify the characteristics of weak intermolecular hydrogen bonding interactions around aromatic systems (Model 6). Visual representations can allow certain attributes in the data to be noticed precisely because they are unusual. A VRML-based model permits us to view different types of data collected together in one scene, and to make links between different levels in the data hierarchy to allow probing of any unusual facets that may emerge in a highly intuitive manner. |
| Model 6. A VRML cluster diagram showing intermolecular hydrogen bonding interactions around aromatic systems |
The model shows the results of a search for short contacts of some typical hydrogen bond acceptor groups to a chlorobenzene ring, normalised against the Connolly surface, i.e. that portion of the van der Waals surface that is accessible to a probe of finite radius. Such an approach makes no presuppositions but rather reveals structural preferences, as is borne out by the fact that the vast majority of the contact vectors are oriented towards the ring hydrogens. The conversion to VRML allows for the vectors, colour coded according to the type of contact group, to be hyperlinked to a second model which in turn can be used to highlight the significant interaction, and give other information such as the unit cell and a literature reference. This reference in turn can be a link to the original electronic journal article which allows the research to progress from the discovery of a potentially interesting effect to reading the original article about it.
Another application of data mining is illustrated in the Panel, which shows how subtle structure-activity phenomena such as hydrogen bonding can be teased out of large amounts of numerical data provided by the technique of molecular modelling.15
|
Adding time-dependence to a model can often enhance the
perception and understanding of a subtle chemical
phenomenon. We experienced this directly when studying
the potential energy surface of a sequence of pericyclic
reactions starting at 1 or 2 and forming
cyclo-octatetraene (4). After a conventional
article had been prepared for printed publication,16 we decided to enhance
it for an electronic version of the journal17 by including selected
3D models, and animating those models to show normal
vibrational modes. Whilst proof-reading the "enhanced"
article, it suddenly dawned on us that the animated mode
for one geometry in particular had unexpected
characteristics.
Ostensibly, the reaction corresponded to two synchronous electrocyclic ring openings proceeding in a particular manner known as conrotation. Each individual ring opening is, according to a theory first proposed by Woodward and Hoffmann, thermally allowed. From another perspective however, the reaction also corresponded to a cyclo elimination reaction thermally forbidden by the Woodward-Hoffmann rules. How could both interpretations be true? Focusing our eyes on the four atoms corresponding to the cyclo elimination, we realised by watching the animation that the Woodward-Hoffmann forbidden characteristics were in fact avoided by a pronounced lateral dislocation of the two reacting centres (Model 7). If you are reading this article in print, and are having difficulty visualising what we mean by that, we strongly urge you to view the animated VRML model to see how we obtained our original insight.18 |
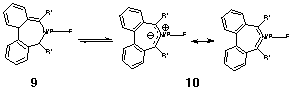
|
| Model 7. An animated model showing vibrational modes. |
Simple animations of this type can also be accomplished using other tools for displaying molecular models such as Chime for molecules or spectra19, but this requires a set of molecular coordinates for every frame of the animation. VRML often requires only the starting and ending coordinates of the molecule. The properties of the various objects within a VRML scene are then interpolated at each time step to create the effect of an animation. Such a technique is particularly effective when animations of large molecules are required.
 |
A quite different application of VRML illustrates how laboratory instrumentation or
apparatus might be depicted (Model 8).20 Having a more "lifelike"
rendition may make it easier for others to reconstruct a
complex experimental set-up. It is also possible to make
the model interactive to facilitate the appraisal of
modifications to the scheme. For example, various VRML
laboratory components or "objects" could be connected
together with the help of a VRML authoring tool from an
on-line "storeroom". The virtual approach would also
provide a means for students to gain some insight to
modern laboratory techniques that might otherwise be
prohibitively expensive to actually carry out.
Ultimately, a network of virtual instruments
corresponding to the layout of real world laboratory
environment could be constructed, and virtual experiments
within the laboratories could correspond to live
experiments.
In the scene shown here, each important object is assigned a "viewpoint" to help in the navigation, and these viewpoints could be used to index the model, and ultimately to search for model components on a global scale. |
| Model 8. A Virtual Molecular Laboratory, with hyperlinks to other information sources. |
There is even the possibility that a real instrument could be depicted and even controlled remotely, in a manner analogous to the automated process controls in a chemical plant. An automated laboratory driven largely by robotics could be managed by an expert from practically anywhere in the world. The clear advantage here is that several such laboratories can be managed by the same person. If the VRML world reflects real world applications, many challenging issues arise. In the case of the virtual laboratory, a mechanism is required whereby the laboratory information is transferred to the VRML scene. If the laboratory information is stored in databases this would entail suitable integration between the VRML model and the database. This can be achieved using a technique known as VRML scripting. Here, little programs are embedded within the VRML model giving it behaviour characteristics. For example, one such program might give a simple VRML model of a clamp the ability to pivot, or in a molecular model, a program might be embedded to calculate a required property of the molecule.