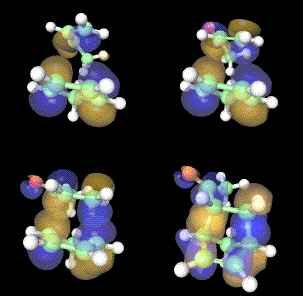
Omer Casher and Henry S. Rzepa
Department of Chemistry,
Imperial College of Science Technology and Medicine,
London, SW7 2AY.
E-mail: o.casher@ic.ac.uk and rzepa@ic.ac.uk
Two IRIS Explorer modules have been developed which fully interact with one or more Mosaic World-Wide Web browsers thereby enabling multiple remote Explorer EyeChem applications to exchange virtual molecular worlds.
During 1994, use of the Internet wide area network for chemical data exchange and collaboration grew very significantly, but has not been matched by corresponding development of appropriate Internet standards and suitable network cognisant tools. From our own research, we have identified two specific areas which we believe urgently require attention. We are addressing the need for Internet based chemical standards by proposing the use of chemical Multipurpose Internet Mail Extension (MIME) content types to transfer semantically rich chemical data[1] via information delivery tools such as World-Wide Web (WWW). Whilst this client-server model is ideal for archiving, we are also convinced of a separate need to develop real-time chemical collaboratories for the exchange of molecular worlds between two or more scientists.
A chemical collaboratory should have four principal attributes. The software should visualize chemical data seamlessly. It should be easily customisable to fulfil the needs of individual research groups. It should be capable of using the Internet effectively and transparently. Finally, it should interface to an archival component such as the World-Wide Web. Because such novel requirements in chemistry had not hitherto been addressed, we undertook the development of EyeChem, a modular visualisation environment (MVE) based on IRIS Explorer. Such modularity and extensibility allowed us to quickly build a pilot system to demonstrate the uses of the high speed UK national network known as SuperJanet.[2] A further advantage is that new molecular data formats can be rapidly and easily assimilated into EyeChem by developing new modules.
A number of EyeChem applications, each dedicated to one particular task, were created by combining new EyeChem and existing Explorer modules. These applications contain a minimum number of widgets and can run without the Explorer GUI showing, thus simplifying the interface. EyeChem modules include file readers to convert files from quantum chemistry data, in Mopac and Gaussian formats, and from crystallographic data into the Explorer Chemistry Pyramid data type. EyeChem Geometry modules convert the Chemistry Pyramid into Explorer geometry data appropriate for ball and stick or ribbon rendering.
In this paper, we introduce a number of new extensions for creating custom EyeChem applications suitable for chemical collaborations and for interfacing with other visualisation systems.
One drawback of an Explorer custom application is that new modules can only be added by reconstructing the application within an Explorer map. This would require the user to have a degree of familiarity with Explorer. To address this issue, we developed the EyeSkm modules to run Explorer scripts. Several separate EyeSkm modules, each containing a frequently used set of commands, can be incorporated within one EyeChem application. Modifying the application, such as attaching a colour map to a geometry module, is carried out by clicking on a button. Any EyeChem application can therefore be custom built to suit the needs of the individual, although the slow resulting performance is clearly an issue that needs remedying. All the EyeChem modules, custom applications and initialising scripts are freely available via an anonymous ftp archive on argon.ch.ic.ac.uk in directory pub/EyeChem.
Eye2eye was our first chemical collaboratory comprising a custom EyeChem application, to which RenderRemote modules are added to export viewing windows with the molecular model to one or more remote collaborators. When picking modules are added and the Render camera locations exchanged between computers, two or more scientists are able to manipulate and edit the same model in real time. In our implementation, explanations were assisted by use of SGI InPerson video conferencing and whiteboarding software. This was demonstrated over a distance between Imperial College and Leeds University using ATM network interfaces and the SuperJanet2 network.
An eye2eye collaboratory has only one computer running EyeChem and remote rendering the molecular model. Remote collaborators are unable to interact with any modules other than the RenderRemote. Ideally, each computer should have its own custom EyeChem session running and a suitable data transport mechanism to interact with the others. By integrating EyeChem with WWW browsers via the Common-Client Interface (CCI) library, we managed to achieve this. The stages of development of this collaboratory are illustrate below with four models.
We had initially developed EyeChem modules to enable visualisation of small molecules and their topological properties such as molecular orbital surfaces. The scientific essence can be captured by creating molecular animation using EyeChem and additional Explorer modules. One such movie illustrating the change in the molecular orbitals involved in a chemical reaction known as Diels-Alder cycloaddition (Figure 1) was saved in MPEG format and allocated a MIME type[3] of video/mpeg on a WWW server. A hyperlink via the so-called Uniform Resource Locator (URL) enables the movie to be viewed using a WWW browser and a suitable MPEG player.[4] This model in effect provides an archival mechanism for a virtual chemical world observed from a particular and unalterable point of view.
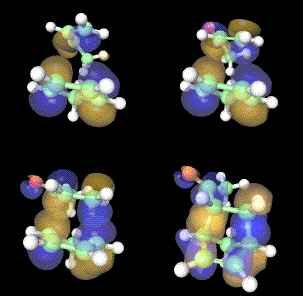
Figure 1. EyeChem application showing four frames in an animation illustrating the frontier orbitals involved in the Diels Alder reaction.
Although a molecular movie conveys more three dimensional information than an image in printed form, the scientist cannot interact with the semantic content of the information. This may be desirable under circumstances when some sense of the three dimensional structure needs to be demonstrated for molecular coordinate files that are not freely available. In general however, semantically rich information is to be preferred. The next stage of the project was to incorporate the semantic content of molecular data in WWW documents.
We chose to use the transport mechanism known as Hypertext transport protocol (HTTP) implemented in WWW servers. Various molecular data files were placed on a WWW server and the HTTP server configured to allocate specific chemical MIME types to the data files via their filename extensions. Hyperlinks to these datafiles were added to WWW pages containing images with their scientific description. A hyperlink would activate a small script resident on the reader's machine. The script initiates an EyeChem session acting on the original molecular data file. The overall effect is that the EyeChem application is serving as a so called WWW "helper" program. This concept of "Hyperactive Molecules"[5] has been incorporated into several of our recent electronically published scientific papers.
One hitherto unsatisfactory feature of the implementation of WWW is the "stateless" nature of the HTTP transport protocol. Very recently, the concept that molecular data files might themselves contain embedded URLs pointing to other relevant information has developed, which goes some way to maintaining "state" between two or more sets of molecular data. Such "self-documenting" molecular data could include references to entities such as hypertext, images, movies and other resources.
Version 2.5 of NCSA Mosaic supports the CCI[6] interface, allowing a symmetrical exchange of URLs and datafiles between Mosaic and external helper programs. We built two new EyeChem modules, EyeCCI and EyeCollaborator, which implement the CCI API. EyeCCI sends a URL to a local Mosaic session while EyeCollaborator sends URLs to one or more remote Mosaic sessions. Once Mosaic has been instructed to "listen" for CCI communication on a specified port number, it will act upon any URLs it receives. EyeCCI can also receive either a URL from Mosaic or the actual data body redirected from Mosaic, and output this as Explorer data to another module.
In this concept of a chemical collaboratory, EyeChem can be used for all required molecular visualisations, whilst other types of content such as hypertext or images can be rendered by Mosaic. Most importantly, EyeChem can interact with Mosaic running remotely. If a remote Mosaic process is itself interacting with another EyeChem session running elsewhere, the two EyeChem sessions are able to interact with each other. Essentially, the CCI/Mosaic combination provides a very fast and cost effective way of implementing the HTTP transport mechanism into EyeChem.
Next we considered how any URL embedded in molecular data might be activated using EyeChem. Most simply, such URLs are identified by parsing the datafile using the Explorer Shell module that runs Unix shell commands or scripts, and then browsing and selecting the URLs with the Explorer BrowseFile module (Figure 2). As each selection is made, the URL can be sent to one or more remote Mosaic or EyeChem windows. Each project could have associated a set of remote collaborator IP host names and CCI port numbers. In this manner, a molecular URL could be exported to e.g an entire classroom of computers, each running their own Mosaic Window and/or EyeChem session. If the URL refers to non-molecular data, it could be directly viewed by e.g. Mosaic, whereas if the URL refers to molecular data as identified by a chemical MIME attribute, then Mosaic could in principle be instructed to retrieve the molecular data using appropriate HTTP protocols, and return it to the EyeChem collaboratory for visualisation.
This mechanism would serve well if only a small number of URLs were associated with any individual molecular data file. In principle however, each individual object such as an atom or a bond could be associated with a URL. For such cases a more sophisticated URL selection mechanism in EyeChem would need to be developed.
If CCI is implemented in non-Explorer applications, there is no reason why communication between EyeChem and these applications could not be achieved. For example, if an AVS equivalent to EyeCCI existed, communication between EyeChem and e.g. the AVS ChemistryViewer via an HTTP engine such as Mosaic would allow the two MVEs to exchange virtual chemical worlds. In this regard, the CCI interface allows genuinely heterogeneous molecular collaboratories to be constructed. The chemical MIME protocol would ensure that such different collaboratories agree on the same molecular datatype definitions.
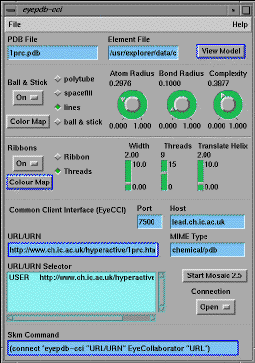
Figure 2. EyeChem Collaboratory displaying URLs parsed from a PDB
molecular data file, and a corresponding Mosaic Window displaying the user
selection.
The implementation of URL exchange via the Common-Client Interface into the EyeChem Collaboratory has several profound implications, not the least of which being that a Virtual Reality Modelling Language (VRML)[7] browser is evolving. The VRML scenario contains multi-participant "virtual worlds" hyperlinked through an appropriate transport mechanism and described using the VRML language, in effect a three dimensional implementation of the World-Wide Web metaphor. Virtual worlds can contain objects hyperlinked to other worlds or to existing WWW based HTML documents. Selection of an object in the complex three dimensional space can in turn be used to invoke further actions. The currently favoured format for VRML 1.0 is a sub-set of the IRIS Inventor file format, which Explorer uses for 3D rendering. If this is accepted as the standard, then extending EyeChem further into a true VRML browser is trivial.
To demonstrate the concept, and to provide a test-bed for VRML developers, we have prepared several sample chemistry-specific VRML[8] files. One illustrates a DNA protein (Figure 3) "anchored" via hyperlinks to two other VRML files, one containing Connolly surface representation of the molecule, and the other containing a ribbon representation of the molecule's protein component. The proposed VRML 1.0, however, does not contain the various surface nodes required for some common molecular representations such as ribbons. All the 3D objects suitable for chemistry but not defined within VRML 1.0 should be defined within a chemistry-specific subset thereof and the VRML browser developed accordingly.
Currently, the EyeChem collaboratory does not fully implement a VRML transport mechanism. It can exchange virtual worlds with collaborators in real time through the remote rendering capabilities of Explorer, as is done with eye2eye. The next step would be to allow datafiles containing VRML mark-ups to be rendered appropriately within an EyeChem collaboratory. In effect, a combination of Mosaic or any other CCI compliant WWW browser and the EyeChem Collaboratory would constitute a full virtual reality system for molecular scientists.
Figure 3. A VRML data file of this DNA protein has been prepared and is viewable by any VRML browser.
An evolving scenario in which chemical collaboratories can be constructed and used via the Internet as virtual reality browsers has been presented. Whilst several aspects of this scenario need further development, the implications of scientists being able to routinely work in this fashion are quite novel and may serve as suitable models for bilateral collaborations between individuals. Hence, the distinction between scientific talks, conferences and scholarly journals may eventually vanish.
We thank Drs. G. A. Suner for valuable contributions, P. Murray-Rust for stimulating discussions and G. D.B. Cameron for the invitation to present this paper on our work.
2 O. Casher, H. S. Rzepa and S. Green, "EyeChem 1.0: A Modular Chemistry Toolkit for Collaborative Molecular Visualisation.", J. Mol. Graphics, 1994, 12, 226. See http://www.ch.ic.ac.uk/jmg/CRG.html
3 N. Borenstein and N. Freed, Internet Request for Comment (RFC) No. 1521 (1993). See also http://www.ncsa.uiuc.edu/SDG/Software/Mosaic/Docs/mailcap.html
4 See H. S. Rzepa and B. J. Whitaker, http://www.ch.ic.ac.uk/chem_mime.html
5 O. Casher, G. Chandramohan, M. Hargreaves, P. Murray-Rust, R. Sayle, H. S. Rzepa and B. J. Whitaker, "Hyperactive Molecules and the World-Wide-Web Information System" J. Chem. Soc., Perkin Trans2, 1995, 7. See http://www.ch.ic.ac.uk/rzepa/RSC/P2/4_05970K.html
6 D. Thompson, "NCSA Mosaic Common Client Interface ", National Center for Supercomputing Applications. See http://www.ncsa.uiuc.edu/SDG/Software/Mosaic/Docs/cci- spec.html
7 G. Bell, A Parisi, M. Pesce, "The Virtual Reality Modeling Language", November 1994. See http://www.eit.com/vrml/vrmlspec.html
8 O. Casher, "A VRML Subset for Molecules", February 1995. See http://www.ch.ic.ac.uk/VRML/vrmlchem.html