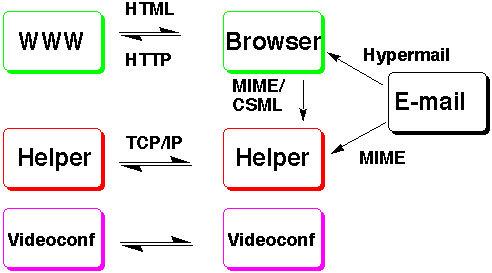
Figure 1. The Interaction between WWW Servers, Browsers, Chemical Helpers and other Information systems.
Department of Chemistry, Imperial College, London, SW7 2AY.
Abstract: We present¶ a "proof-of-concept" model of an Internet based chemical Collaboratory. This is based on an integration of a World-Wide Web server running the HTTP protocol, hypertext-markup language based browsers, molecular visualisers based on Explorer EyeChem modules and browsers implementing the Virtual-reality modelling language 3D scene description.
A parallel development in the use of Internet based networks in the last two years has been the remarkable growth of the World-Wide Web (WWW), a hypertext based global information system. In the last year in particular, a number of innovative applications in the area of chemistry have been introduced,[2] including a concept termed "hyperactive" molecules[3] in which a set of molecular coordinates stored on a WWW server can symbolically hyperlink into a WWW document. This document is retrieved from the server using a protocol known as HTTP (Hypertext-transfer-protocol) and displayed on a so called WWW browser using markup commands known as HTML (hypertext-markup-language). Using a further mechanism known as MIME[4] which has been adapted for chemical applications,[5] the WWW browser, which is itself incapable of rendering the molecular coordinates, can pass these onto a chemical "helper" for visualisation. Such a helper can be specified by the local user to be e.g. an appropriate EyeChem module, although in principle any suitable program can be selected. The logic of the communication between the various components, together with the protocols used is shown in figure 1.

Figure 1. The Interaction between WWW Servers, Browsers, Chemical Helpers and other Information systems.
From the aspect of designing a Collaboratory involving two-way communication, the mono-directional MIME protocol operating between a document browser and the molecular visualisation system is a severe limitation. In particular, it is not possible for any form of hyperlink which might be present in a molecular data file to be acted upon appropriately. The molecule file is an information cul-de-sac. Whereas hyperlinks in a document composed of HTML can reference molecular data, the reverse is not true. Because HTML is basically a two dimensional page description language and molecular data is often intrinsically three dimensional, efficient information flow between the two different types of data file is essential. In this paper, we present one possible solution to this problem, and introduce an alternative and we think complementary approach of having three dimensional molecular "scene" descriptions defined using a newly introduced protocol known as VRML, or virtual-reality modelling language.
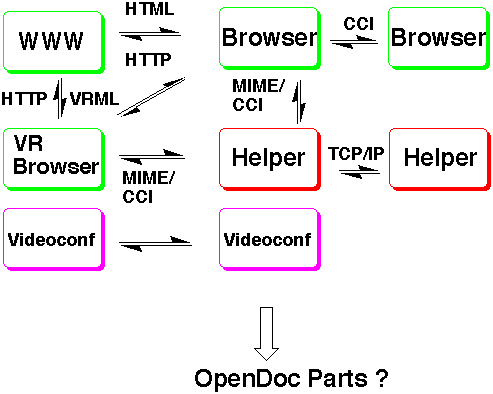
Figure 2. An WWW Server/WWW Browser/EyeChem/VRML Interaction achieved using CCI Communication.
This combination of modules means that URLs or the future URNs (Uniform Resource Names) can now be designed into various molecular datafile formats. The one we selected to illustrate the point is the protein databank (pdb) format, which can be extended with USER commands to include URL citations. Such molecular URLs could be associated with the entire molecular datafile, with specific regions of the molecule(s), or perhaps their properties, or simply with individual atoms or bonds. Such URLs could point to other network based resources such as other molecular datafiles, images, databases, text or any useful and relevant collection of information. In effect, the molecular datafile becomes self-annotating, and one might even imagine that an entire scholarly paper could consist of a single molecular datafile with appropriate URLs pointing to discussion of the various features of interest. When visualised using EyeChem, resources which cannot be rendered are simply passed on to Mosaic or indeed other appropriate programs for viewing. One can even imagine a heterogeneous molecular visualisation system, where for example the Explorer based EyeChem and the AVS based Chemistry viewer could collaborate, each processing the same data in their own way. Also enabled now are remote network based collaborations between one user employing EyeChem and another using AVS. In effect, we are now achieving a true chemical Collaboratory.
The metaphor of a Collaboratory can in fact be extended further into non-molecular "worlds". Explorer rendering is based on use of the Iris Inventor 3D object library and the recently proposed VRML 1.0 (virtual reality modelling language)[7] in fact uses a sub-set of this library. Inclusion of EyeCCI makes EyeChem therefore equivalent in functionality to a VRML browser, albeit one particularly attuned to rendering molecular 3D objects. We note with interest that several commercial VRML browsers are under development, including a product called WebSpace which is also based on Iris Inventor. This latter product can also communicate with WWW servers, via another WWW browser called Netscape. Thus the combination of EyeChem/Mosaic/CCI is partially equivalent to WebSpace/Netscape. The latter does not presently use the CCI protocol for internal communication, but an alternative. The WebSpace/Netscape product does not in fact compete directly with the EyeChem system, simply because VRML 1.0 is currently not well suited for the particular needs of representing molecular objects.[8] Thus constructs such as tapered bonds, dot surfaces, ribbon representations etc are beyond the current scope of VRML. We approach this problem by placing a URL pointing to the actual PDB file within a VRML file. EyeVRML will send this URL to EyeCCI.This module in turn will retrieve the file and pass it through standard EyeChem modules for handling the graphics not supported by VRML 1.0. By contrast, the WebSpace approach is for the VRML viewer to support a superset of the VRML 1.0 nodes, basically all Inventor nodes. The disadvantage of this approach is that any VRML file written for WebSpace might not be viewable by other VRML viewers.
We have used EyeChem to produce a number of molecular scenes in VRML format, available from a WWW server for immediate viewing using a suitable VRML browser.[9] The advantages of having such a combination is that one might expect the development of efficient and fast VRML browsers for a variety of computing platforms, which will achieve the true inter-operability necessary for collaboratories. Another advantage of establishing links with the VRML community is to achieve continuity with other non-molecular "scene" descriptions.
[2] H. S. Rzepa, B. J. Whitaker and M. J. Winter, J. Chem. Soc., Chem. Commun., 1994, 1907.
[3] O. Casher, G. Chandramohan, M. Hargreaves, P. Murray-Rust, R. Sayle, H. S. Rzepa and B. J. Whitaker, J. Chem. Soc., Perkin Trans 2, 1995, 7.
[4] N. Borenstein and N. Freed, Internet Request for Comment (RFC) No. 1521.
[5] H. S. Rzepa, P. Murray-Rust and B. J. Whitaker, Internet Draft, March-August, 1995, rzepa-chemical-mime-types-01.txt.
[6] D. Thompson, The Common Client Interface, NCSA, October, 1994. See http://www.ncsa.uiuc.edu/SDG/Software/Mosaic/Docs/cci-spec.html for further details.
[7] G. Bell, A. Parisi and M. Pesce, VRML 1.0; A proposed specification for a Virtual Reality Browsing system. See the URL http://www.eit.com/vrml/
[8] O. Casher, "A VRML Subset for Molecules", http://www.ch.ic.ac.uk/VRML/vrmlchem.html
[9] O. Casher and H. S. Rzepa; http://www.ch.ic.ac.uk/VRML/.