|
Definition of Chemoinformatics: The
collection, representation and organisation of chemical
data to create chemical information, to which theories
and models can be applied to create chemical knowledge.
Objectives of these lectures: To
introduce the background to
the course, and the skills to be acquired during
the course laboratories, including the use of computer
workstations, computer software and network information
resources and prioritising and organising the
information obtained using these tools. To introduce
the chemistry computer laboratory sessions and what you
are expected to do during these sessions. The lecture
summaries below contain numbered items. At the end of
the course, you will be expected to be familiar with
the techniques and concepts outlined in each of these
29 categories, and be able to apply them to researching
a project.
This course does not deal with any
aspects of data logging, analysis and mining (often
called Chemometrics) e.g. Excel spreadsheets,
Mathematica, MatLab etc.
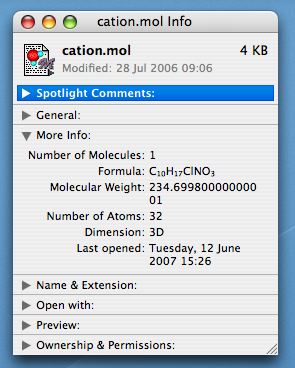 Data: Managed by
Operating Systems (OS) on
Computers (Windows XP, Mac OS X, Redhat Linux), on Phones
(e.g. Symbian, Win CE, OS X), Organisers (e.g. PalmOS), iPods (OS X) Data: Managed by
Operating Systems (OS) on
Computers (Windows XP, Mac OS X, Redhat Linux), on Phones
(e.g. Symbian, Win CE, OS X), Organisers (e.g. PalmOS), iPods (OS X)- Access: On computers by
authentication against User names/passwords.
Some users (root,admin) have special
permissions.
-
Organisation: Data normally held
in Files located in Hierarchical Folders
(Directories). Directories referred to as
Home or My
documents have special status for
each user.
- Files: naming convention
uses 8.3 (DOS) vs 31 (MacOS 9) vs 256 (Modern)
characters. Allowable filename characters based
on ASCII set with some exclusions (space, $, /,
:, ? etc). Some OS Filenames are case
sensitive (Unix), others are not
(Windows). Often the cause of much
confusion!
- File Content/Data type:
normally indicated by adding a 2-4 character
extension after a period (.doc) to the name. The
extension may or may not be
visible. Special types, used by the
OS, may be invisible by virtue of starting with a
period. The (free text) file content may have
been indexed and hence is searchable by the OS.
- File Metadata (Properties):
Creation/Modification Dates, sizes, access
permissions, "ownership", content, etc is
organised by the OS. ( "the ability to look at your
hard drive through a metadata-filtered
view"
- File Location in hierarchy
performed by searches using file metadata as
criteria.
- File Size: In "bytes"
(approximately, 1 character = 1 byte, sometimes 2
bytes). 106 bytes =~1 Mbyte,
109 bytes = ~1 Gbyte, 1012
bytes = ~1 Tbyte. Maximum size for any file
normally 2 Gbyte (Windows) or very much larger
(Linux, Mac OS X).
- Archives: A collection of
Folders and Files which preserves the hierarchy
and file metadata (.zip, .sit, .tar).
-
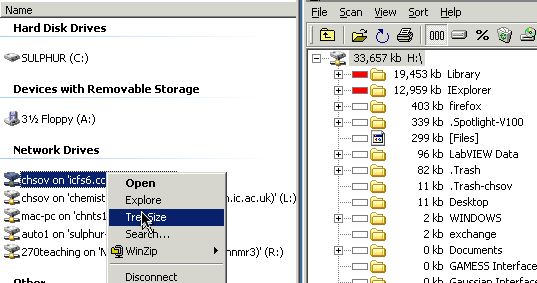 Storage: Storage:
-
Permanent Data Storage, as
files on:
-
Local
hard drives (capacity 40 Gbytes to 750
Gbytes)
-
Network Drives:
- Home directory (Desktop icon
Home, also known as drive
H:\, capacity ~200 Mbytes per
user)
- Drive L: (Your "Home" on Linux
systems)
- Drive N: (A data-silo)
- Drive R: (Where files from
departmental NMR Spectrometers are
placed)
- Removable media (PenDrives, iPods,
CD-RW/DVD, capacity 128Mbyte - 16Gbyte - 120 Gbyte)
-
Temporary Data Storage, as
- "clipboard" in "System Memory" (capacity
not known by user, but probably < 10
Mbyte)
- cache or temporary files, not normally
seen by the user but can wreak havoc if
corrupt!
-
File Usage: Data Files are created
and exchanged using:
- Combinations of programs, typically a
Word processor (Word), a chemical drawing program
(Chemdraw) and
Bibliographic database (EndNote).
- Data exchange between these programs
using copy/paste via clipboards or via
files (drag-n-drop,
save/open).
-
File Data Structures: Internal
structure of files can be hidden or
exposed.
- Hidden (binary) file (or
clipboard) formats are normally understood only
by specific programs. Examples include .DOC, .RTF
(Rich Text format), .GIF, .PNG, .JPEG (Graphics),
.MPEG (audio, video), .PDF (Acrobat).
- Exposed structures include
HTML (structured Hypertext markup language), SVG (Scalable Vector graphics),
TXT (un or semi-structured text)
-
Chemical types include:
- Molecule specifications, with atom
connection co-ordinate types such as SMILES,
PDB,
Molfile
- Spectral/analytical specifications such
as JCAMP
- Query specifications such as SD
-
Data: Semantics (meaning) can
be added to data structures to make it
re-usable in different contexts: XML
(eXtensible markup language) is the best known
way of doing this.
- MetaData: Data should have
descriptions to add context. HTML can have
exposed metadata (i.e. this document). Acrobat
has structure for metadata (XMP) but this is
rarely used!
-
Data Transport:
-
Using Wires/Fibres
- Local to computer: USB2 (480 Mbps), Firewire (800 Mbps),
internal workings.
- Between Computers: Ethernet (up to 1
Gbps)
-
Wireless
- Local to computer: Bluetooth (e.g.
keyboards, mice, phone, ~1 Mbps)
- Between Computers: WiFi (Chemistry
library, labs, lecture theatres, ~40 Mbps)
-
Data Exchange: Human/Computer
Interactions (i.e. human specifies search
query, computer responds with an answer)
-
Session-centric (i.e. the
context between the query and the answer is
preserved during the session, 1977-present)
- Exchange of General Graphics using
X-Windows (e.g. eXceed), Citrix
(proprietary), Windows Remote Desktop
(Windows).
- Exchange of Chemistry graphics: Beilstein
Commander, SciFinder.
- Exchange of Programs and Services: .Net
or
Java
- Real-time:
MOO/Chat/IRC/AIM programs, Whiteboards,
Games, Realtime media Streaming/Broadcast,
videoconferencing.
- Document-centric
(1993-present) via Web Browsers via URLs, HTML
and MIME/chemical
MIME
- Information Object-centric
(1997-present) Web Browsers, NewsFeeds/Podcasts
using RSS/XML.
- Data Exchange:
Computer<=>Computer/Human
Interactions: The
Semantic Web; A Trusted semantic web: Digital
Certificates.
- ⇒ Coursework
|
 Data: Managed by
Operating Systems (OS) on
Computers (Windows XP, Mac OS X, Redhat Linux), on Phones
(e.g. Symbian, Win CE, OS X), Organisers (e.g. PalmOS), iPods (OS X)
Data: Managed by
Operating Systems (OS) on
Computers (Windows XP, Mac OS X, Redhat Linux), on Phones
(e.g. Symbian, Win CE, OS X), Organisers (e.g. PalmOS), iPods (OS X) Storage:
Storage:
![[BACK]](BackBtn.gif)
![[NEXT]](NextBtn.gif)