| [Molecules: None] [Related articles/posters: None] |
The traditional role of a conference scientific panel is to structure the content into themes, based in part on an awareness of the current state of research in the area, and in part in the expectation that authors will produce material corresponding to these themes. Because traditional conferences then run along what might be described as a non-digital route, there is little opportunity to analyse the structures and themes that actually develop in practice.
Operating a conference using the Web as a medium provides an opportunity to analyse the proceedings using a variety of statistical and analytical tools which are available for the purpose, and in particular to applly chemical similarity algorithms to the database of content that represents the conference.1
The conference started as a collection of 120 individual articles. These articles were only related to each other by the fact that belong to the conference and that they had been allocated places in themes. The chemical information was extracted from the articles and used for similarity and clustering analysis. The results of this poster have been integrated into the conference as lists of related articles and posters on the top of each article.
Capturing the chemical information from the conference was not trivial. Most of the information was in the reaction schemes. These were originally drawn by the authors using their chemical drawing packages and then converted to the GIF format for publishing on the Internet. A call was made to the authors to supply their original chemical drawings, of which some authors did do. It was expected that it would be easy to inter convert these drawings with other molecular file formats, but this was not the case. The art work contains many chemical abbreviations and generic abbreviations, for example COOH, which the chemical programs usually resolved into methyl groups in the interconversion.
The rest of the GIF images were passed through a chemical OCR program to convert the bitmap image back to the original chemical drawing for further conversion to a molecular file format. Unfortunately the OCR programs were designed to work on images of 300 dpi or greater rather than the low 72 dpi resolution of the Internet. This approach was abandoned, since the structural error rate meant it was almost always faster to redraw the structure.
There were some articles that had molecules in molecular file formats from x-ray crystallographic co-ordinates. Unfortunately there were only 104 of these molecules, a very small proportion of the total number of molecules from the conference. The rest of the molecules had to be redrawn so that the required chemical information was available. Only 1,300 key molecules were drawn out of the possible 5,000 in the conference, the remainder being simple analogues which were not entered. For example, such information was often contained as table lists, and the association of an R group with its structure could not be performed automatically.
Comparing the articles were done by using fingerprints of the molecules and the articles themselves. A fingerprint is a series of bits that represent the information of an object by which bits are set on or off. The molecules were individually encoded using the algorithms in the Daylight Chemical Information Systems, Inc. toolkit. The molecules were also encoded into fingerprints using two other algorithms. One was based on the number and sizes of the rings in the molecules and the other was using a fragment dictionary where each bit represented the existence of a fragment from all the possible fragments generated from the captured molecules in the conference. Each article was also encoded so that each word was converted to a pseudo molecule before applying the Daylight toolkit algorithms.
Each article had many fingerprints generated from the words and molecules. These fingerprints needed to be combined together so that it was possible to compare different articles with each other. All the fingerprints from an encoding algorithm were summed together so that each bit position no longer represented the existence of a feature, but the number of molecules that had that bit set for that feature in the fingerprint. This was termed a paperprint. In Figure 2 is an example of ten fingerprints, of length 24 bits, added together.
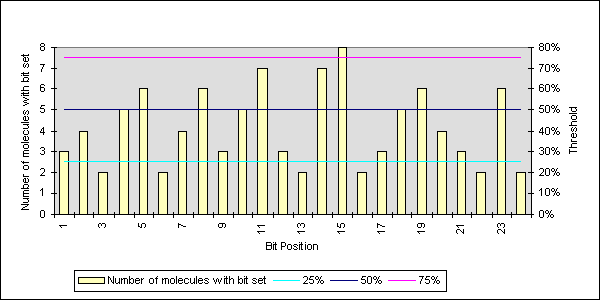
The method used for finding the similarity coefficient between two fingerprints, was optimised to look at bit streams. The paperprint was normalised back to bits, by looking at the bits greater than a threshold. It was found that a threshold of 50% was sufficient for similarity analysis. It was important that the number of bits set was enough to create reasonable similarity coefficients. Too many bits set will cause saturation, indicating that all the articles were similar to each other. The opposite effect occurred when not enough bits were set in the fingerprints. Unfortunately a lot of information was lost during the normalisation stage.
The similarity coefficients, using the Tanimoto coefficient, between papers were low, around 0.2, which showed that the articles were not very similar using these algorithms. Instead the ranking of articles relative to each other was used. In the headers of the articles the ranking of each article was a combination of the molecular, ring descriptions, fragments and the text encodings. The ranks from each of these methods were added together and the whole ranking system was re-normalised so that the ranks were back to 1st, 2nd etc..
Displaying data in graphs is the easiest method to see trends. Using a toolkit called AGLO 2, concept maps were drawn from the ranks. The nearest 1 and 5 neighbours were drawn. AGLO was designed to produce graphs that maximises some measure of desirability, in its case to produce pretty graphs. It considered connections between nodes, or papers, to be equal and iterated them to produce a graph so that the vertex and edge distances are maximised/minimised to satisfy the constraints imposed. The constraints were chosen to produce clusters and strings of articles. Strings were valuable since they provided routes through the conference. Each step would go to a similar theme, and eventually completely different disciplines would be reached.
| Nearest neighbour | Nearest 5 neighbours | |
|---|---|---|
| 2D |  | 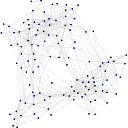 |
| 3D | 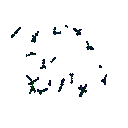 |  |
To view the 3D VRML files, you will need your browser to be configured using an appropriate e.g. Netscape plug-in such as Live3D or CosmoPlayer.
The nearest neighbour plots show the articles in discrete clusters. The distance between the clusters has no relation to the differences in the clusters nor do the intra-cluster distances have any relation to the absolute similarity in the articles. Both the 2D and 3D plots are active, thus clicking on the points in the graph will go to the relevant article. The nearest 5 neighbours plots, the ranked articles mentioned in the article headers, were more clustered around three major sections with articles bridging these sections. There were a couple of articles in the centre of this plot which were quite close together due to the surrounding article connections, but were not connected to each other.
Unfortunately there is no way of integrating the actual similarity coefficients into the AGLO plots. The distances between articles were totally dependent on the number of connections between them.
If the chemical information was readily available at the beginning of the conference then the navigation tools could be created for participants use before the conference started. The availability of the chemical structures would have made it possible to set up e.g a Daylight database ready for structural similarity searching, a valuable tool to accompany the existing keyword systems.
These methods have clustered the articles. Unfortunately it is very difficult to relate these clusters with human clustering methods, for example the conference themes. The algorithms need to be changed so that they mimic the methods humans use to decide which articles belong together. The same experiment will be performed on the chemical content on the associated conference ECTOC-1 to see whether there are any differences due to the conference theme.