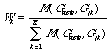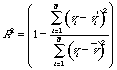Molecular Graph Metric Selection With The Help Of Genetic Algorithm.
Yurchenko Ò.À.1, Kumskov
M.I.2, Vorona D.G.3
1 Moscow State University, Calculated Mathematic and
Cybernetic Faculty;
119899, Russia, Moscow, Vorob'evy Gory, MSU.
2 Zelinsky Institutes of Organic Chemistry, Russian Academy
of Science;
117913, Moscow, Leninskii prospect, 47, Mathematical Chemistry Lab.,
Fax: (095) 135-53-28, E-mail: kumskov@cacr.ioc.ac.ru
3 Moscow State University, Mechanic and Mathematic
Faculty;
119899, Russia, Moscow, Vorob'evy Gory, MSU.
Abstract. The selection technique of molecular graph's metrics on the
baso of the genetic algorithm is offered. The metrics are defined at feature's
space of large dimension, where a descriptor is number of fragment's repetition
in given molecular graph. Two variants of metric selection criterion are
considered: first - on the basis of a Shannon's information measure, second -
on the non-parametric estimation of molecular properties.
Introduction. The problem of properties forecasting of chemical
substance can be considered as the special case of a problem of image
recognition [1] with the teacher, in which the object (molecular graph in this
case) should be described by a vector of attributes. One way of such
representation uses "structural spectrum" of a molecule: the list of all
fragments of given complexity, meeting in set, is formed and the repetition
number of each fragment in molecular graph gets out as attribute. Such
approach has been well tested, because it provides fast automatic calculation
on graphs and provides universality, i.e. the opportunity to use it as a
description for wide chemical class
Statement of a problem. Let we have training set of substances
(or molecular graphs)), at which the values of some property is known. We
define complexity of fragments and then determine a set of descriptors, which
will participate in the metric formation. We form a matrix "molecule -
attribute" X= (Xij), i=1,..., N; j=1,...,
M; where Xij - a number of repetition of j-th fragment in
the i-th molecule, N-number of molecules, M-number of
descriptors (fragments). Further with the help of genetic algorithm [2,3] there
conducted the selection of various variants of the metric. On the basis of each
variant it will be carried out the cluster analysis of objects (for example,
with the help of k-average cluster algorithm) , then received
decomposition is estimated to its adequacy to provide the classification of
molecules on the given property. On each step the best metrics (i.e. metrics,
inducing best decompositions) are selected which will be participate in
generation of new metric's variants.
Genetic algorithm. "Gen", which determined the current decision
of a problem, is represented by a vector w[[propersubset]]R
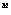 ,
where w
,
where w
 is the weight factor of j-th descriptor in the given metric. On the
initial stage random family of the gens (decisions) is formed, which contain
Q gens. For each member of family ( for each parent) new metrics (the
children) are formed. Received childrens are selected and Q "best" ones
(according to selection criterion) form the next generation of decisions.
is the weight factor of j-th descriptor in the given metric. On the
initial stage random family of the gens (decisions) is formed, which contain
Q gens. For each member of family ( for each parent) new metrics (the
children) are formed. Received childrens are selected and Q "best" ones
(according to selection criterion) form the next generation of decisions.
Genetic operations: (1) "Mutation": change the parameters of the
parent in one gen's component (in our case this is the change of one of the
weight factors):
w
 (k+1)=
w
(k+1)=
w
 (k)
+ Δ w , where k - the number of a current
step. The mutation frequency and their size can get out by various ways [2,3].
(k)
+ Δ w , where k - the number of a current
step. The mutation frequency and their size can get out by various ways [2,3].
"Crossover": for two given parents two children are formed, which
contained a gen part from one parent, and another part - from other. For
example, we can generate a random binary vector
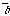 =(b
=(b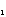 ,...,b
,...,b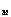 );
b
);
b
 [[propersubset]]
{0,1}; j=1,...,M . Then the first child receive j-th weight
of the first parent, if bj =1, and weight of the second parent, if bj
=0; and the second child recieve complementary values . The choice of the
parents can also be various:
[[propersubset]]
{0,1}; j=1,...,M . Then the first child receive j-th weight
of the first parent, if bj =1, and weight of the second parent, if bj
=0; and the second child recieve complementary values . The choice of the
parents can also be various:
1. All Q parents are divided into pairs, and each pair forms two
children, thus we receive Q crossover's children from current
population;
2. All possible parental pairs (Q * (Q-1) /2 pairs) are considered which
generate Q * (Q-1) children.
Selection criterion. The selection can performed: (à) only among
children; (b) among children and parents. The second variant is more
preferable, since the found optimum decision-parent will remain in all
selections. We shall consider two variants of selection criterion.
Shannon's information measure. With the help of given property
one can divides all molecules as "active" and "inactive". Then one can
perform the cluster analisis of objects. Let we get k clusters, which
contain gi graphs (i=1,...,K), .where
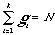 Let the i-th cluster have ai "active" and bi of "inactive"
molecules, where ai + bi = gi . Then we can define the "quality" of
i-th cluster as the function
Let the i-th cluster have ai "active" and bi of "inactive"
molecules, where ai + bi = gi . Then we can define the "quality" of
i-th cluster as the function
Ôi = -pi *log(pi ) - qi
*log(qi ), where pi = ai /gi ;
qi=bi/gi
The quality criterion of total cluster decompositin we can define as
Ô(k)=
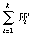 *Ô
*Ô where
weight factor Wi = gi /N.
where
weight factor Wi = gi /N.
This function Ô(k) is the selection criterion. It is clear, that
Ô(k) characterizes a heterogeneous degree of the clusters (ideal
variant - when we receive two equal clusters, and one has only "active", and
another -only "inactive" molecules; in this case Ô (k) =0). The
minimization of this criterion within the framework of genetic algorithm leads
to generation of metrics, that the molecules of one class are located
compactly.
Non parametric property estimation [4]. We shall define the
"similarity" of two molecular graphs G1 and G2, as,
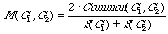
where Common (G1, G2) is the number of common fragments in G1 and
G2, S(G) is a-number of all fragments in graph G.
Let training set contain N molecular graphs {G1,G2,...,GN} with
known meanings(importance) of investigated property v(Gi). For the
performing property estimation of new graphs Gnew we calculate K
most similar graphs {Gj1,Gj2,Gj3... GjK}. An estimation of property
v for graph Gnew will be calculate under the following formula:
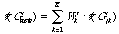 where
where
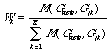
As Gnew consider consiquently all graphs of training set and build a
error vector of an estimation of properties v*=(v*(G1),
v*(G2),...,v*(GN)).
Let define as the selection quality criterion of the metric the multiple
correlation coefficient R of a buld error v*:
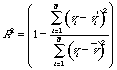
The described approach is under realization for IBM PC 486. It is planned to
conduct research of a genetic algorithm behaviour for classification both
physico-chemical, and biological properties of substances.
This study was supported by the Russian Foundation for Basic Research
(project nos. 96-01-01598 and 94-01-00041)
References
1. Gorelik A.L., Skripkin V.A. Methods of recognition / 2-th edition,
Moscow, Higher School, 1984, 208p.
2. Davis L. Handbook of genetic algorithms / Van Nostrand, Reinhold, New
York, 1991.
3. Fogel D.B. Applying evolutionary programming to selected control
problems. / Comput. Math. Applic., 1994, vol.27, no.11, p.89-104.
4. Kumskov M.I., Mitushev D.F. Expert metric formation for recognition of
the molecular-chemical graphs. Non-parametric estimation of the substance's
property / Abstracts of 2-nd All-Russia Conf. "Pattern recognition and
Image Analysis, Russia, Ul'janovsk,1995, part 3, p.135-136.
Return to main WATOC Poster page
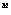 ,
where w
,
where w
 is the weight factor of j-th descriptor in the given metric. On the
initial stage random family of the gens (decisions) is formed, which contain
Q gens. For each member of family ( for each parent) new metrics (the
children) are formed. Received childrens are selected and Q "best" ones
(according to selection criterion) form the next generation of decisions.
is the weight factor of j-th descriptor in the given metric. On the
initial stage random family of the gens (decisions) is formed, which contain
Q gens. For each member of family ( for each parent) new metrics (the
children) are formed. Received childrens are selected and Q "best" ones
(according to selection criterion) form the next generation of decisions. (k+1)=
w
(k+1)=
w
 (k)
+ Δ w , where k - the number of a current
step. The mutation frequency and their size can get out by various ways [2,3].
(k)
+ Δ w , where k - the number of a current
step. The mutation frequency and their size can get out by various ways [2,3].
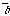 =(b
=(b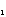 ,...,b
,...,b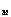 );
b
);
b
 [[propersubset]]
{0,1}; j=1,...,M . Then the first child receive j-th weight
of the first parent, if bj =1, and weight of the second parent, if bj
=0; and the second child recieve complementary values . The choice of the
parents can also be various:
[[propersubset]]
{0,1}; j=1,...,M . Then the first child receive j-th weight
of the first parent, if bj =1, and weight of the second parent, if bj
=0; and the second child recieve complementary values . The choice of the
parents can also be various: 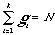 Let the i-th cluster have ai "active" and bi of "inactive"
molecules, where ai + bi = gi . Then we can define the "quality" of
i-th cluster as the function
Let the i-th cluster have ai "active" and bi of "inactive"
molecules, where ai + bi = gi . Then we can define the "quality" of
i-th cluster as the function 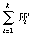 *Ô
*Ô where
weight factor Wi = gi /N.
where
weight factor Wi = gi /N.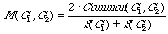
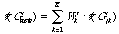 where
where