
As a part of a project directed to the design and synthesis of novel inhibitors for collagen biosynthesis, we focused our attention on conformationally restricted Pro-Gly dipeptides of type 2. The use of these heterocyclic units as scaffolding devices in Pro-Gly peptide sequences 1 should lead to the stabilization of the secondary structure.

A considerable number of methods have been developed for the preparation of 2-oxazolines and 2-thiazolines which are present in many biologically active natural products[4]. However, a general synthetic method to introduce chiral oxazolines and thiazolines in peptide structures has to be developed.
The synthesis of the heterocyclic systems 5 is outlined in Scheme 1. The cyclization of the imino ether 4 and (R)-cysteine or (L)-serine alkyl ester hydrochlorides provided an efficient and simple route to 5[5] in good yields (60 - 80%). The coupling was affected in anhydrous EtOH at 60C being complete after 8 hours. The key intermediate 4 is easily prepared from N-tert-butoxycarbonyl-(S)-proline via a two-step procedure involving transformation of 3 into the corresponding amide, followed by alkylation of the amide with Meerwein salt.
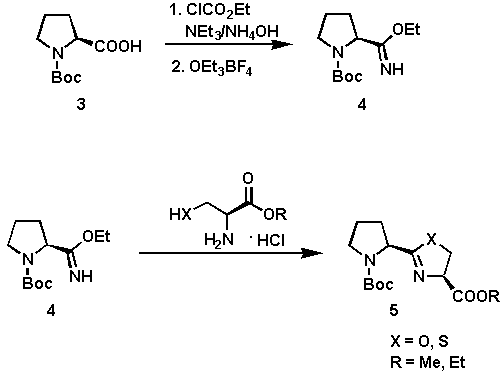
Scheme 1
The development of compounds designed to mimic certain secondary structural features of peptides is an important approach for elucidating the receptor bound conformation of a biologically active peptide. One major secondary structural feature of many biologically active peptides is the beta-turn which is characterized by its phi2, psi2, phi3, and psi3 torsion angles (6, Figure 1).
It has been proposed that prolyl 4-hydroxylase which catalyzes the formation of 4-hydroxyproline in procollagen by the hydroxylation of proline residues, recognizes a type-II beta-turn conformation formed at the X-Pro-Gly segments in nascent procollagen chains[6,7]. In an ideal type-II beta-turn, these torsion angles possess values of -60, 120, 80 and 0, respectively[8,9]. Several conformational constraints have been designed to mimic an ideal type II beta-turn and one or two of the torsion angles[10,11] are generally restricted. Our designed system 2 simultaneously restricts two (phi2, phi3) of the four torsion angles as shown in Figure 1.
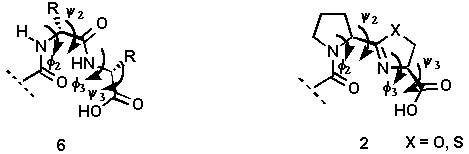
Figure1. A comparison of the structural features of a beta-turn 6 with those of the designed mimic 2.
Computational studies were undertaken to evaluate the ability of the designed system 2 to adopt the desired type-II beta-turn conformation. A full systematic conformational Monte Carlo search was performed upon the N-acetyl-N'-methylamide model compounds 7 and 8 by examining all combinations of rotable torsion angles[12]. Two force fields (MM2 and Amber) were investigated and the calculations were carried out using both gas-phase minimization and the GB/SA continuum solvation model[13]. The results are summarized in Table 1. The calculations using the Amber all-atom force field have not yielded significantly different beta-turn structures.
Table 1. Torsion angles and relative energies of minimized conformations of 7 and 8 using gas-phase minimization (method A) and the GB/SA solvation water model (method B).
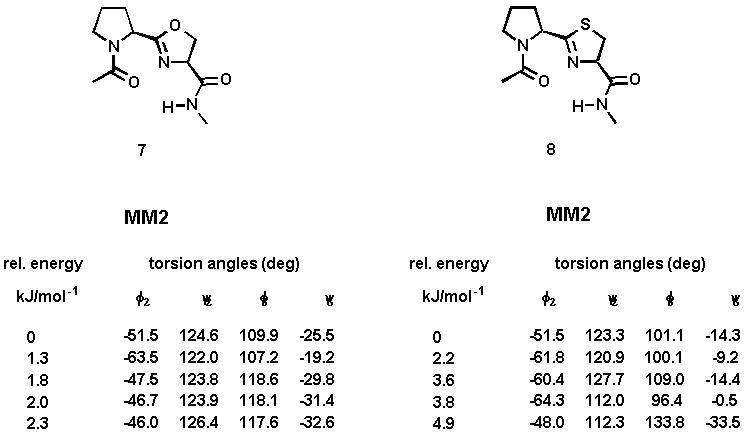
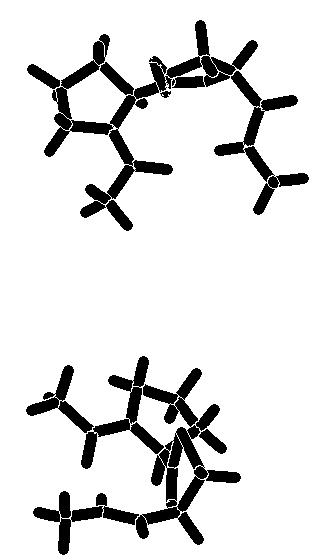
The calculated torsion angles phi2, psi2, phi3, and psi3 are in good agreement with those of the classical type-II beta-turn. The turn is stabilized by an intramolecular hydrogen bond between the carboxamide hydrogen and the prolyl N-carbonyl oxygen. The distance between these hetero atoms is 2.8 - 3.0 Å (for 7 and 8, both calculations), a value well within the range observed for hydrogen-bonded beta-turns in peptides[14]. An additional hydrogen bond between the carboxamide hydrogen and the nitrogen atom of the heterocyclic ring (distance: 2.4 Å) contributes further to the stabilization of the beta turn. A graphical representation of the energy-minimized structures of 7 and 8 is enclosed. Superimposition of the global minimum of 7 and 8 and the ideal type-II beta-turn revealed an RMS deviation of the main chain atoms of 0.341Å (7) and 0.298 Å (8), indicating that a high degree of similarity existed in the turn region. The comparison was made by fitting nine atoms of the backbone of 7 and 8 (calculation method A) with the corresponding amide backbone atoms of the classical type-II beta-turn.
Graphical representation of the energy-minimized structures of
7 and 8.
This study contributes a simple and direct approach to peptidomimetic compounds. Modeling studies have shown that the conformation of the designed system 2 is in close agreement with that of the hydrogen-bonded type-II beta-turn with respect to the torsion angles and the interatomic distances. We believe that these systems can serve as useful conformational constraints which, when incorporated into the structures of selective bioactive peptides, will yield new conformationally constrained peptide analogues for structure activity relationship studies. Incorporation of the peptide-mimicking building block into biologically relevant peptide structures is in progress.
Further details of the modeling studies and the experimental data of the synthesized compounds are available upon request. I gratefully acknowledge the support by FibroGen, Inc.
1. Gante, J. Angew. Chem. Int. Ed. Engl. 1994, 33, 1699 - 1720.
2. Giannis, A., Kolter, Th. Angew. Chem. Int. Ed. Engl. 1993, 32, 1244 - 1267.
3. Spatola, A. F. In Chemistry and Biochemistry of Amino Acids, Peptides, and Proteins, Ed. Dekker, M., Vol. 7, pp. 267 - 357), New York, 1983.
4. Lockhart, M. I., Newton, G. G. F., Abraham, E. P. Nature 1954, 173, 536.
5. North, M., Pattenden, G. Tetrahedron 1990, 46, 8267 - 8290.
6. Günzler, V., Brocks, D., Henke, St., Myllylä, R., Geiger, R., Kivirikko, K. I. J. Biol. Chem. 1988, 263, 19498 - 19504.
7. Harding, J. J., Crabbe, M. J. C., Post-Translational Modifications of Proteins, CRC Press, London, 1992.
8. Smith, J. A., Pease, L. G. Crit. Rev. Biochem. 1980, 8, 315 - 399.
9. Ball, J. B., Hughes, R. A. Tetrahedron 1993, 49, 3467 - 3478.
10. Genin, M. J., Ojala, W. H., Gleason, W. B., Johnson, R. L. J. Org. Chem. 1993, 58, 2334 - 2337.
11. Freidinger, R. M., Veber, D. F., Hirschmann, R., Paege, L. M. Int. J. Pept. Protein Res. 1980, 16, 464 - 470.
12. Chang, G., Guida, W. C., Still, W. C. J. Am. Chem. Soc. 1989, 111, 4379 - 4386.
13. Still, W. C., Tempczyk, A., Hawley, R. C., Hendrickson, T. J. Am. Chem. Soc. 1990, 112, 6127 - 6129.
14. Taylor, R., Kennard, O. Acc. Chem. Res. 1984, 17, 320.