Towards the Chemical Semantic Web. An introduction to
RSS.
Peter Murray-Rusta and Henry S.
Rzepab
aUnilever Centre for Molecular Informatics,
University of Cambridge, UK, bDepartment of
Chemistry, Imperial College London, SW7 2AY.
Abstract: We describe the essential features of a
meta-data based news and alerting service known as RSS, and
call for its adoption within the chemical community. We
describe how RSS could be used as one of the essential
components for constructing a chemical semantic Web.
Keywords: RSS (RDF Site Summary), XML (eXtensible
Markup language), CML (Chemical Markup language), Semantic Web,
News alerts.
It is appropriate after almost exactly ten years of
Web-based publishing to assess what this revolution has meant
for the dissemination of scientific and chemical information.
Well in excess of 2 billion documents, of which perhaps 5% have
some scientific content, have been generated (authored is
perhaps putting it a trifle too strongly). To this one must add
a limitless number of "virtual" documents generated dynamically
upon request. Even within a relatively specialised discipline
such as chemistry, the effect upon a human can be overwhelming;
keeping up with the literature has evolved from a weekly browse
of the tables of content of perhaps ten key paper-based
journals to having to visit a much larger number of
computer-based Web sites. It might be fair to say that many
humans are not coping too well! Ironically of course, computers
are much more efficient than humans at scouring a large number
of information sources in an error, boredom-free and periodic
manner; the challenge is merely in specifying what it is they
should be on the lookout for. The first generation Web
(circa 1993-present) turns out in retrospect to have
been rather unsuited to this task. This is not actually the
fault of the original design, merely a facet of how it was (or
more accurately was not) implemented. The key omission was
meta-data, this being a concise and structured declaration of
the content model of the document.
Before this last topic is elaborated, its worth noting the
"Google phenomenon", an index of the entire Web which for most
scientists renders it actually useful as a scientific
information source.1 Thus the erstwhile need to
constantly "bookmark" key locations found by a possibly
accidental and probably not reproducible path, is now
productively replaced by being able to rely on Google to find
information based on a few choice key words. Google augments
this with additional meta-information derived from choices made
by earlier searchers using the same key words; in effect a
coarse but gradually accumulating peer review mechanism which
naturally selects the survival of the fittest information.
The original and still current Web, Google not withstanding,
in practice contains very little overtly structured information
in the form of declared meta-data; this may be simply the title
of a document and little else. The human authors of most
documents have either cared little or knew little about the
relatively arcane mechanisms the original designers of e.g HTML
had put in place to capture meta-data. This is something of a
chicken and the egg issue; the average chemist has hitherto had
no compelling reason to learn these arcane methods, preferring
instead to focus on chemical applications and doing what they
do best! The consequence has been that the Web has not
fulfilled its anticipated potential for "serendipity", or the
art of accidental and fortuitous discovery of unexpected
information. This is still an act expected to be achieved by
humans exercising their perceptive skills and not by
computers.
At this point in the discourse we introduce RDF-based Site
Summary or RSS (acronyms can indeed be successful at capturing
the world's imagination, viz HTML!). This is a simple
but powerful XML-based implementation of meta-data of which
(like HTML) the arcane aspects can be hidden behind
user-friendly software referred to variously as a news reader,
aggregator or RSS client. RSS grew out of an idiosyncratic
phenomenon known as Web logging. Web Logs (now known as
"Blogs") are essentially personalized Web servers containing
chronologically organised items of information. Four items of
meta-data are implicit and indeed mandatory in a Web Log; the
identity of the author, the date of each item, a brief
description of it and how to link to it. The "blogging"
community perceived the need to summarise Blogs using such
meta-data and then to broadcast these summaries in a manner
which allowed "aggregation" of themes into larger "channels" of
such information, and thence into syndication; a procedure not
very different from e.g. newspapers or broadcast
television. This process in due course merged with another and
rather more formal vision of evolving the homogeneous
unstructured Web into a Semantic Web,3 where in
addition to meta-data, information is carried which allows
machines to process documents without the necessity of human
intervention. The resulting fusion has resulted in a formal
XML-based specification known as RSS2, which is
supported by ready availability of RSS-aware software which can
act upon the contents of such RSS files. The remainder of this
article shows an example of how RSS is currently applied and
used, and follows with a call for its application in chemistry
and a scenario of how it might be applied in this subject in
the future.
Creating RSS
One way of regarding RSS is as relating to the management
and description of hyperlinks in the same way that the earlier
HTML (and more recently XML) markup languages relate to the
management and declaration of content and data. RSS therefore
augments a conventional Web site (or the smaller scale Web
log), and can be added to a site without the need for any
additional infrastructures or software. Many major news sites
already do this, but it is still rare for chemically related
sites. At the server end, deploying RSS requires nothing more
than the addition to the contents of a Web site of two
entries;
- the creation of a document (by default variously named
index.rss or rss.xml) which defines meta-data for a channel
of related documents and links. The example shown in Scheme 1
follows the W3C-supported specification known as RSS
1.02, which includes support for RDF (Resource
Description Framework), the latter being described as a
"lightweight ontology system for supporting the exchange of
knowledge on the Web".2 We note here a competing
(and simpler) XML-based specification currently known as RSS
2.0 (here the acronym stands for Really Simple Syndication)
2 which does not include the RDF components; both
are recognised by most current RSS software (but not all the
components are necessarily processed).2 We
recommend here the use of RSS 1.0, since this has the greater
potential for extension into domains such as chemistry, but
the choice is not critical since the two formats can always
be easily interconverted in the future using e.g. XSLT
stylesheets.
-
In order to assist the robot-based "auto-discovery" of this
file, an entry can (optionally) be made to the root
document of the Web server (i.e index.html or equivalent)
identifying the location of the RSS document
<link rel="alternate" type="application/rss+xml" title="RSS" href="http://.../index.rss" />
In fact, the RSS document need not even be real; it can
(perhaps even should be) generated dynamically by appropriate
query of any content management system which is used to
generate documents for the Web site. Like HTML, RSS can also be
generated using simple authoring tools; a good one is to be
found at http://rssxpress.ukoln.ac.uk/. An example
of an RSS file is shown in Scheme 1:
<?xml version="1.0"?>
<rdf:RDF
xmlns:rdf="http://www.w3.org/1999/02/22-rdf-syntax-ns#"
xmlns="http://purl.org/rss/1.0/"
xmlns:dc="http://purl.org/dc/elements/1.1/"
xmlns:cml="http://www.xml-cml.org/schema/cml2/core" >
<channel rdf:about="http://www.xml-cml.org/">
<title>Chemical Markup Language</title>
<link>http://www.xml-cml.org/index.html</link>
<description>CML Highlights</description>
<webMaster>admin@cmlconsulting.com</webMaster>
<image rdf:resource="http://ww.xml-cml.org/cml.gif" />
<items>
<rdf:Seq>
<rdf:li rdf:resource="http://www.xml-cml.org/rss/" />
</rdf:Seq>
</items>
</channel>
<item rdf:about="http://www.xml-cml.org/rss/">
<title>This RSS file contains an embedded Molecule in CML</title>
<link>http://www.xml-cml.org/cml.rss</link>
<description>Currently, RSS clients are not capable of acting
upon the cml namespace in the RSS descriptor, and so ignore this
information. In future, one can anticipate that a CML-aware RSS client
will not only extract the molecular information, but be capable of
searching/filtering/transforming it into more useful forms (ultimately
for example being able to detect particular sub structures or other
specific items of interest to the reader).
</description>
<dc:creator>Henry Rzepa</dc:creator>
<dc:date>2003-04-06T10:00:00-05:00</dc:date>
<cml:molecule id="a1" title="water">
<cml:formula conciseForm="H 2 O 1"/>
</cml:molecule>
</item>
</rdf:RDF>
Scheme 1. A representative RSS file using the RSS 1.0
specification. Note particularly that although the CML
component is not currently processed by any standard RSS
software, its presence does not disable processing of the other
components by such clients.
Using RSS
The design of RSS includes one subtle concept which takes it
beyond the conventional use of a language such HTML. Whereas
the latter is really intended to be presented to a human in an
easily read and hence visible form, achieved of course by using
a Web browser, RSS (in common with all XML languages) is
intended to be offered to both humans (via the
equivalent of a Web browser) or to software and other agents
for more automated processing.
-
The best way of viewing an RSS feed or channel is to
install an appropriate client and add the URL of the RSS
file on the remote server. Some RSS clients can also
"auto-discover" such files. Invoking http://purl.org/net/syndication/subscribe/?rss=http://www.xml-cml.org/index.rss
for example will initiate the following process.
- The RSS file http://www.xml-cml.org/index.rss is
passed to a syndication service.
- This service checks for the existence of the
file
- The RSS file is then validated to ensure it conforms
to an appropriate RSS specification.
- A page listing a number of current RSS clients is
presented. The user can then install an appropriate
one4.
- The same page allows the RSS channel to be subscribed
to the just installed RSS client
-
Typically, the user will install a relatively controlled
and limited list of RSS channels which they find are
particularly relevant to their interests (Figure 1).
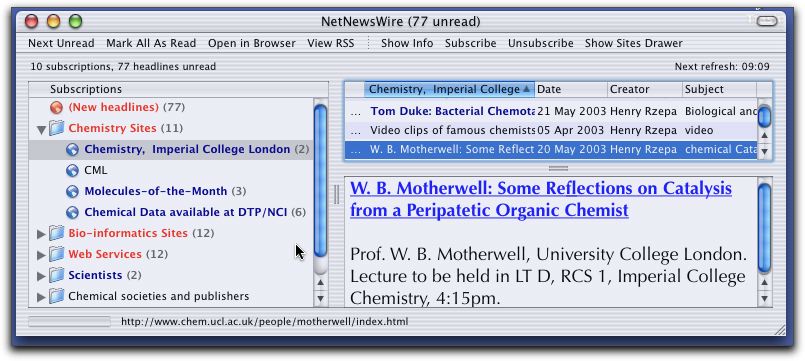
Figure 1. Typical User interface to an RSS client.
This can be done either by selecting from pre-determined
lists provided with the RSS client (although "science" as a
category is included with most clients, this currently
contains little of overt chemical interest), by discovery
at visited web sites (RSS feeds are normally indicated by
displaying the following icon:  ) or by searches at sites specializing in RSS
discovery5. We emphasize that numerous RSS
channels already exist; it might be expected that in due
course, syndication sites specializing in chemical content
will arise.
) or by searches at sites specializing in RSS
discovery5. We emphasize that numerous RSS
channels already exist; it might be expected that in due
course, syndication sites specializing in chemical content
will arise.
-
The RSS client will then periodically perform various
automatic tasks.
- Foremost, it will periodically revisit each RSS site
to check if any new information or items have been added.
An interesting variation is an RSS client that converts
any new items to an alerting email.6
- The retrieved meta-data will normally be indexed by
the RSS client to enable facile local searches of this
content to be performed. Some clients can also archive it
to the user's hard disk for longer term
availability.
- The items retrieved will be aggregated, sorted by
various meta-data attributes, such as author, date,
subject etc and presented in a condensed form for rapid
browsing by the user (Figure 1).
- Although not yet a reality, several RSS clients
promise more sophisticated filtering of aggregated items.
Thus the user may indicate that only items by a
particular author are displayed, or those that contain a
particular keyword or subject. This process can be
arbitrarily sophisticated. Thus (as exemplified in the
RSS example shown in the box) the meta-data could consist
of molecular definitions carried using the appropriate
XML descriptor known as CML,7 and the user
could specify that a chemical sub-structure comparison be
carried out on the RSS feed such that only items that
reference e.g. a heteroaromatic ring containing
two nitrogen atoms be displayed. Importantly, this action
is now performed by software, and need not rely on the
perceptive capabilities of a trained human chemist. We
note that it is not possible to perform such checks using
a conventional Web browser, and conventional HTML!
-
It is not necessary that the RSS file be processed by a
visually-oriented RSS client. It is entirely feasible that
other types of automatic software actions can be set up. A
relatively simple one which illustrates the potential of
this approach is invoked by http://rssxpress.ukoln.ac.uk/view.cgi?rss_url=http://www.ch.ic.ac.uk/index.rss
This passes the contents of the RSS file to (in this
instance) an XML process which applies a (XSLT) stylesheet
to transform the RSS content to a different form (in this
case a convention HTML-based web page, but arbitrarily any
other XML form). A variation on this procedure is to
include an expansion of an RSS channel within an existing
document, to in effect update the document automatically
with relevant information, e.g. the addition of
<script
src="http://rssxpress.ukoln.ac.uk/lite/viewer/?rss=http://www.ch.ic.ac.uk/index.rss"></script>
to an HTML document.
The results of such requests could then be re-used in a
different context, as for example automated entry to a
database, use by a synthesis robot, entry in a personal
diary etc. One might imagine an example in the future where
RSS channels from primary chemical journals will contain
explicit chemical information encoded in e.g.
CML7 which are automatically screened by the
user's software for the presence of say a particular
molecular structure, or molecules with particular
properties. Any found matching these criteria could be
automatically submitted for e.g. quantum mechanical
calculation of further properties.8 The human
comes to their (probably virtual) desk in the morning to
find not only that overnight the system has discovered a
molecule of interest to them, but has arranged calculation
of its properties, or even its synthesis in the
laboratory!
Current applications of RSS.
Clients for simple processing of RSS have been available since
around 2000. A site specialising in indexing and searching only
RSS-enabled documents9 estimates that by mid 2003
some 34,000 instances containing references to around 1 million
documents existed; a search using the term "chemistry" however
revealed little significant content (yet). Some such do exist;
we list the following two as indicators of the potential.
-
MDB - The Metalloprotein Database and Browser at the
Scripps Institute.10 The URL for the RSS file
takes the form:
http://metallo.scripps.edu/services/api.php?func=metallopdb&metal=zn&count=12&format=rss
This in fact is a dynamically generated RSS feed,
resulting for a query fed to the (MySQL) database of
metalloproteins, with the output being formatted in RSS.
Installing this will alert the user to any new (in this
case Zinc containing) metalloproteins recently added.
-
The National Cancer Institutes support an RSS
feed11 of the form:
http://dtp.nci.nih.gov/dtpstandard/servlet/chemRSS.
This is an feed generated using a Java servlet
application to again query a database and return an
appropriately formatted RSS file.
Even simpler implementations
deploy RSS to e.g. alert the community to new additions to our
local "molecules of the month" column and to the subjects of talks to be
presented by visiting seminar
speakers (Figure 1).
Adoption within Chemistry of RSS and the Creation of a
Chemical Semantic Web.
As noted above, the chemical community makes little current use
of such news feeds; this article is a call to rectify this
situation! Whilst some of the (chemical) examples described
above might be considered fanciful in 2003, we emphasize that
other applications can created immediately.
An obvious and immediate application might be as an alerting
service for the primary chemical journals. Consider the current
somewhat haphazard procedure most chemists have come to adopt
since the majority of science journals have gone on-line. One
periodically finds a browser "bookmark" to a favorite journal
(and anticipating it is still functional) a visit to the
"latest issue" section of the table of contents will reveal
titles of the latest articles, and possibly an abstract that
may impart more explicit chemical information (but only
visually since it is almost certainly a graphical image which
requires a human for perception). Further analysis will require
the download of e.g. an Acrobat file, which normally
arrives on the user's disk in the "downloads" directory. It
still requires much action by the user to organise this reprint
into a bibliographic database (for example Endnote), and the
process has to be repeated for each journal, with the added
difficulty that each publishers' "user-interface" is different
and has again to be learnt. Most of us probably realise that
this procedure does not "scale", and are wondering how we will
cope with this in the future. This procedure could be replaced
by a much more structured one based on RSS-derived metadata,
derived (in the future) by automated processing of the original
full (XML-based) article. In order to discover new and
potentially interesting articles, the user subscribes to the
RSS feeds of relevant publishers, and can e.g. simply
search the latest items that appear automatically for key words
of interest. The article download is still necessary, although
it may be possible for the RSS client to automatically invoke
e.g bibliographic software (or alternatively such software
could support RSS directly). When primary scientific
publications become available directly in XML (rather than
e.g. Acrobat) the possibilities for their re-use
increase enormously; no longer is one limited merely to
printing the article!
Another immediate application of RSS is as an alerting
services for new additions to chemical data bases (although
here the sheer volume of new additions might require immediate
mechanisms for filtering this down to a manageable quantity).
In addition to the two examples noted above, we have easily
implemented a simple extension to our php/MySQL-based ChemStock
inventory system12 to alert users to e.g the
last five added entries at any given time. It is also apparent that the strength of
the system is that separate alert streams from different
communities (say chemistry and bio-informatics) might be
semantically combined to create connections that are simply not
being currently made due to the sheer overload of information.
Perhaps the most significant aspect of an increasing deployment
of RSS is that it could serve as a focal point for increasing
awareness of the importance of creating properly structured
information, which includes well defined meta-data, and of
managing the links between such information (the hyperlinks) in
a manner which allows software as well as humans to utilise
these connections in a semantically meaningful manner. RSS does
seem to be a tool which is bringing the ultimate vision of a
chemical semantic web one step closer.3
References
- Google: http://www.google.com/
- For the formal specification, see http://web.resource.org/rss/1.0/. For the
RSS 2.0 specification, see http://backend.userland.com/rss An RSS based
channel for tracking development of W3C recommendations is http://www.w3.org/2000/08/w3c-synd/home.rss.
More information can be found at http://www.w3.org/2001/10/glance/doc/howto#rss
- P. Murray-Rust, and H. S. Rzepa "Towards the Chemical
Semantic Web", Proc. 2002 International Chemical Information
Conference, ed H. Collier, (Infonortics), 2002, pp 127-139;
T. Berners-Lee and J. Hendler, "Publishing on the semantic
web", Nature, 2001, 410(6832), 1023-4.
�
- At the time of writing, we found http://www.awasu.com/ for Windows operating
systems, or http://ranchero.com/netnewswire/ for MacOSX
work effectively.
- RSS syndication services include http://www.syndic8.com/ or http://rssxpress.ukoln.ac.uk/ for the
UK.
- For RSS2email conversions, see http://www.w3.org/2002/09/rss2email/
- P. Murray-Rust and H. S. Rzepa, "Chemical Markup, XML and
the Worldwide Web. Part 4. CML Schema", J. Chem. Inf.
Comp. Sci., 2003, 43(3), in press and references
therein.
- For further discussion of extended CML families such as
CMLComp, see P. Murray-Rust and H. S. Rzepa, chapter in
"Handbook on Chemoinformatics", Ed J. Gasteiger and T. Engel,
Wiley, 2003, in press.
- See http://www.rss-search.com/
- J. M. Castagnetto, S. W. Hennessy, E. D. Getzoff, J. A.
Tainer, M. E. Pique and E. Michael, "Data sharing in
Bioinformatics: Using the MDB's web services.", Abstracts of
Papers, 225th ACS National Meeting, New Orleans, LA, United
States, March 23-27, 2003, COMP-141.
- D. Zaharevitz, personal communication.
- H. S. Rzepa and M. J. Williamson, Internet J.
Chem., 2002, article 6.

 ) or by searches at sites specializing in RSS
discovery5. We emphasize that numerous RSS
channels already exist; it might be expected that in due
course, syndication sites specializing in chemical content
will arise.
) or by searches at sites specializing in RSS
discovery5. We emphasize that numerous RSS
channels already exist; it might be expected that in due
course, syndication sites specializing in chemical content
will arise.