XML in Chemistry
Peter Murray-Rusta
and Henry Rzepab
aUnilever Centre for Molecular
Informatics, Cambridge University
bDepartment of Chemistry, Imperial
College of Science, Technology and Medicine.
1. Markup Languages and the publication process
Although the use of markup languages in publishing goes back to
the 1960s and e.g. the introduction by IBM of GML (Generalised
markup language) and its subsequent evolution into the SGML
standard, most authors are nowadays more familiar with one
particular more recent implementation referred to as HTML
(Hypertext markup language). The rapid impact of its use in
conjunction with the World Wide Web was in large measure due to
its ease of use to achieve presentational and visual effect;
its limitations as a mechanism for expressing precisely defined
data and meanings were not always adequately recognised. These
limitations meant that in areas such as molecular sciences
where precise meanings are essential, a variety of often
proprietary solutions continued to be used to define and
manipulate molecular "data" and information. The publishing
processes were seen as quite separate, and the process of
translating data, information and knowledge into a published
entity remained an activity requiring much human perception. It
is also worth noting that the reverse process of converting the
published materials back in usable data remained equally human
intensive and hence expensive.
The need to reconcile these two extremes was recognised at
the first World Wide Web conference in 1994, and this gelled
shortly after in a remarkable communal effort towards the
specification of an extensible markup language called XML, with
the ultimate vision of what has been described by Berners-Lee
as the creation of a "Semantic Web".1 The objectives
of this impressive effort included the following;
- To provide a more universal infrastructure for
publishing
- A recognition that the use of XML will require subject
specific vocabularies ("ontologies"). An ontology is defined
as a description, such as a formal specification of a
program, of the concepts and relationships that can exist for
a (software) agent or a community of agents.
- To provide a mechanism for enhancing quality
("validation")
- To promote the creation of dynamic hyperdocuments
- A recognition of the need to be able to re-use components
of documents for other purposes
- To provide a mechanism for creating "smart archives", in
which the re-usable components (information objects) can be
readily identified
- To create an infrastructure for underpinning the emerging
areas of e-business
The vision included therefore the creation of a new generation
of ontologically rich primary publication, and a clear division
of the respective roles of humans and software agents (robots).
Thus humans should be able to:
- publish all their data automatically
- eliminate "errors" from publications
- use the published literature as a database
- "understand" information from other domains
Robots should be able to:
- analyse publications (on whatever scale)
- create secondary publications
- purchase chemicals
- synthesize chemicals from literature
To achieve this, we argue that a number of prerequisites must
be in place.
- Automatic data capture, especially from instruments. We
note here that we have moved from a generation some 30 years
ago where data capture from instruments was often only
analogue (chart paper), to the use of standard computers to
capture and process data, to most recently an increasing
tendency to place these computers on-line and connect them to
centralised data stores.
- Common ontologies for the molecular science
community.
- Ontologically guided authoring
2. An example of the issues involved in "Capturing"
Chemistry
The following extract2 from a typical molecule
science journal illustrates both how precisely data and
information must be represented, but also how much human
perception is required to translate this information as
presented in this (linear) form into e.g. a reproducible
experiment or a mechanistic interpretation;
"Thiamin phosphate synthase catalyzes the formation of
thiamin phosphate from 4-amino-5-
(hydroxymethyl)-2-methylpyrimidine pyrophosphate and
5-(hydroxyethyl)-4-methylthiazole phosphate. The reaction
involves... dissociative mechanism...carbenium ion
intermediate...and pyrimidine iminemethide observed in the
crystal..."
Note the profusion of chemical structure information,
concepts and terms, which only a trained human chemist could
easily process. Quantitative concepts and units are also
ubiquitous;
"A 500 ul aliquot of 0.8 uM TP synthase in 50 mM Tris-HCl
(pH 7.5) and 6 mM MgCl2 incubated at room
temperature with 50uM CF3HMP-PP."
An even greater degree of human perception is required when
handling graphical chemical representations, which may contain
many, often fuzzy and dangerous, human-only semantics (e.g. 2D
representations of 3D properties, relative stereochemistry,
aromaticity, hydrogen and other "weak" bonding, use of generic
and "R" groups, reaction arrows and mechanisms, etc etc). The
challenge therefore is to develop an infrastructure which can
be routinely used to capture, store and appropriately filter
and display such information.
3. The Current Position of XML (2002)
We will argue here that XML offers a general powerful and
extensible mechanism for handling both the "capture" and the
publication of chemical information, and most particularly for
the first time will allow this process to operate equally well
in both directions. Our basis for stating this derives from the
following observations:
- XML is increasingly widely accepted as an information
infrastructure
- The protocols are all public and many of the tools
open(source).
- XML is vendor-neutral, but with heavy vendor
involvement
- There is a large communal investment in generic
tools (e.g. business2Business, e-commerce)
- XML has a modular approach; an application is built from
components
- The expectation that domains will create domain-specific
XML protocols and tools
- XML is increasingly universal in backends, middleware,
servers
- XML has a rapidly increasing support from database
vendors
- XML has close interoperability with other informatic
standards: (UML, OMG/CORBA, etc.)
- There is increasing support for "XML over the net" and
browsers (e.g Internet Explorer, Netscape 6, etc).
- XML is very well supported by books, tutorials.
4. Global Open Activity in Scientific XML
So how has the scientific community adopted these concepts? As
noted above, the first World-Wide Web conference in 1994
specifically identified Maths and Chemistry as requiring
specific markup. The first WWW conference provided the spark,
and during the period 1995-7, CML (chemical markup language)
evolved to become the first XML language, and a concurrent
effort lead to MathML becoming formalised as such in
1998.3 We estimate that by 2002, perhaps 50
specifically scientific applications has been described in some
degree (for example 37 are quoted on one XML
portal4, the Science Citation Index shows around 570
references to the keyword XML and SciFinder retrieves 38
references on the concept "XML in chemistry").
We also emphasize that XML is designed to allow markup
languages to be combined, at whatever level of granularity, and
hence documents could contain any number of components deriving
from specific XML languages. HTML, which we noted above, has
evolved into one such language (XHTML) but in its latest
development, has been modularised into smaller, more easily
implemented components (for example, XFORMS, a data entry and
validation component can be implemented separately from other,
more display oriented components), and XHTML can co-exist in a
document with e.g. SVG (a scalable vector graphical language),
MathML and CML. We elaborate this when discussing namespaces
(vide infra).5
5. Some Essentials of an XML system
The tasks that can be identified in implementing an XML
solution include;
- The creation of documents from both legacy sources of
data and de novo by humans
- The creation and capture of metadata (dictionaries of
terms, tables of contents, codes, etc.)
- Specification of namespaces (a reserved addressing scheme
for information)
- Human validation of the system (conformance to agreed
specifications)
- Machine validation of documents (according to a specified
and agreed schema)
- Document transformation (XSLT)
- Rendering and display (XSL-FO, Domain-specific such as
molecular representations)
The design of an XML-based markup language should provide
for;
- a simple, extensible DTD or Schema (do not over
complicate, and make it modular)
- Agreed semantics
- One (or more) agreed and published ontologies
- Agreed examples and conformance tests
- A community of critical mass
Appropriate tools for accomplishing this should be identified.
These might include;
- XML Writers
- XML Readers (more difficult than readers since the XML
may not be normalised to a single form)
- Legacy converters (difficult because of variation and
ambiguity in the original data which may require some degree
of perception for an accurate conversion)
- Validators
- Dictionaries
- Editors
Custom written XSLT stylesheets and generic editors will do
some of these, but a DOM (Document object model, which
represents a syntax free abstraction of the data in memory) is
probably essential for many subjects
6. Ontologies of relevance to Chemistry
An overview of the types of ontologies required is shown in
Table 1.
| General
Non-chemical informatics |
Business and Commerce, Government,
Regulatory, Academic, Publishing, etc. |
Reuse existing or emerging
approaches |
| Domain-specific
Non-chemical |
Mathematics (MathML), Healthcare (HL7/XML),
Genomics (GeneOntology), etc. |
Collaborate to reuse existing or
emerging approaches |
| Chemical specific
but generic information types |
| Numeric data, Descriptive prose, Safety |
Ontologies must be created by the
chemical community, reuse generic tools |
| Chemical-specific
information types |
Chemical substances, Molecules, Analytical and
Spectroscopic,
Reactions, Computational chemistry, |
The Chemical community must build
the complete tool set |
Of the chemically-specific information types, support should
include that for;
- Molecules and substances
- Reactions
- Analytical information, especially spectra
- Computation and simulation (QM, mechanics, dynamics,
etc.)
- "Data-centric" concepts (numbers, units, arrays,
matrices, etc.)
- Specialist software for display, editing, searching
etc
- Support "adjoining" disciplines such as bio areas,
materials science etc.)
7. XML DTDs and Schemas
In this section, we outline some of the existing generic tools
and protocols for creating valid XML documents.
The DTD (Document type definition) is a concept rooted in
SGML, and is still used in XML to constrain the Markup
vocabulary (i.e. the basic elements used for markup) and to
some extent the (sub)structure of documents (i.e. what element
can be a parent or child of another).
Schemas are a more recent development, and unlike DTDs, are
themselves expressed using XML. Of particular relevance to
chemistry, they provide advantages over DTDs in that they can
also be used for;
- Datatyping: numbers and user-defined types
- enumeration (for example to specify the list of chemical
elements)
- Lexical patterns
- Inheritance
- To allow additional user-created rules
(Schematron/XSLT)
Schemas and dictionaries also support:
- Conversion to software (e.g. CML-DOM)
- Authoring support (e.g. in editors)
- Data validation on entry
The use of DTSs, and Schemas in particular, for creation of
valid documents can bring enormous benefits, including
eliminating/reducing software failure due to the use of
invalid data and reducing difficulty of (human)
understanding due to invalid publications.
8. Namespaces
Each information object must be uniquely named to avoid
collision and ambiguity. This is achieved using XML
namespacing.
-
The example below shows a paragraph of text (derived from
XHTML, which inherits the default namespace), within which
components of CML are embedded with prefixes using the
defined namespaces;
<html
xmlns="http://www.w3.org/1999/xhtml"
xmlns:cml="http://www.xml-cml.org/schema/cml2/core">
<p>We can supply the following set of molecules:</p>
<ul>
<li><cml:molecule id="p1" title="phosphine">
<cml:atomArray>
<cml:atom elementType="P" hydrogenCount="3"/>
</cml:atomArray>
</li>
<li><cml:molecule id="p2" title="penguinone"/></li>
</ul>
</html>
-
The next example illustrates how CML can be used in
conjunction with the STMML namespace6 to specify
units and their constraints:
<molecule id="m1">
<crystal spacegroup="Fm3m" z="4">
<stm:scalar title="a" errorValue="0.001" units="angstrom">5.628</stm:scalar>
<stm:scalar title="b" errorValue="0.001" units="angstrom">5.628</stm:scalar>
<stm:scalar title="c" errorValue="0.001" units="angstrom">5.628</stm:scalar>
<stm:scalar title="alpha" errorValue="0">90</stm:scalar>
<stm:scalar title="beta" errorValue="0">90</stm:scalar>
<stm:scalar title="gamma" errorValue="0">90</stm:scalar>
</crystal>
<atomArray>
<atom id="a1" elementType="Na" formalCharge="1" xyzFract="0.0 0.0 0.0" xy2="+23.2 -21.0"/>
<atom id="a2" elementType="Cl" formalCharge="-1" xyzFract="0.5 0.0 0.0"/>
</atomArray>
</molecule>
- STMML is a proposal6 for domain-independent
components for Scientific-Technical-Medical information, and
contains key elements such as Units,
Dictionary, Metadata, item,
array, matrix and supports Datatypes
such as numbers, max/min, ranges, errors, etc.
- A more extended example of this concatenation of
namespaces7 contains up to eight namespaced
components, and illustrates how a complete publication in
XML/CML could be achieved.
The use of namespaces can be seen in a more general context
in Figure 1, which illustrates how the various specific XML
components might relate to each other.
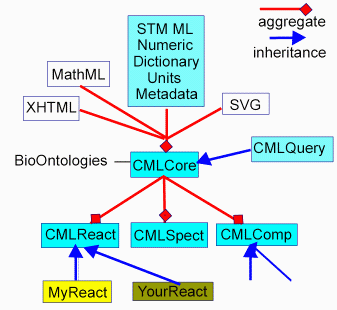
In particular we note here how the original CML
specification8 can be extended by modularisation
into a Core namespace, and extended via other schemas into
e.g.
- CMLReact. A reaction, containing
reactantLists, productLists and links
between them.
- CMLComp. A container for computational and
simulation input and results
- CMLQuery. A generic query language
- Hooks for other Schemas such as e.g. SpectHook,
for spectral parameters and data, and links to molecular
details (assignment)
9. Dictionaries and Schemas
It is useful to separate the domain ontology from the
Schema/DTD, which allows the schema to be more abstract and
which helps extensibility. Thus a 3- or 4-level hierarchy can
be envisaged:
- The data instance
- The XMLSchema describing the instance
- The dictionary/ies describing the instance
- The schema describing the dictionaries
where an instance document refers to NAMESPACED dictionaries to
add semantics and ontology. In this system, units are
themselves verified by the UNITS dictionary. An overview of
this process is shown in Figure 2.
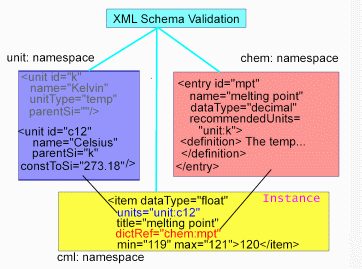
Structure of Dictionaries
We summarise briefly below some characteristic features of
dictionaries:
- Dictionaries consist of curated entries, and
Many dictionaries are "flat" with seeAlso, e.g. the
IUPAC GoldBook
-
A Single hierarchy is common:
- generic ("isA"):
eukaryote <-- vertebrate <-- mammal <--
human
- partitive ("hasA"):
body <-- leg <-- foot <-- toe
- Dictionaries can now be namespaced for uniquification and
navigation
- Dictionaries must have curatorial information
- Dictionaries should support versioning
The existing IUPAC dictionaries provide a natural base for
creating an XML-based machine processible resources. These
dictionaries fall into three broad categories; Descriptive
(e.g. Medicinal Chemistry, Phys. Org. Chem., Stereochemistry,
etc), validating (e.g. Theoretical Chemistry) and supplemental
(e.g. Atomic Weights).9 Their availability for
XML-based processes would be a considerable asset.
10. XML and Metadata
Metadata is an important component of a document or information
object, and it can serve a number of purposes:
- Navigational/Discovery. How is a piece of
information to be discovered, e.g. e.g. Dublin Core and
GILS
- Descriptive. What does the information mean and
how is it to be used?
- Constraining. What constraints are there on the
structure and content of the information. Is it
valid?. this would be accomplished using mainly
XMLSchemas.
- Supplementary. Additional (hyper)data added from
metadata
- Algorithmic. Deductions can be made from metadata,
using e.g. Schematron and XSLT and RDF.
- Chemical-descriptive. e.g. Medicinal, PhysOrgChem,
GoldBook, StereoChem
- Chemical-constraining. e.g. Theoretical Chemistry,
CIF
- Chemical-supplemental. e.g. tables of Atomic
Weights, dictionaries of compounds etc.
- Chemical-algorithmic. TheoChem, CIF
Communally agreed schemas for defining such metadata are again
seen as an essential component of the XML-infrastructures.
11. Conclusions
In this brief review of the application of XML in chemistry, we
have summarised the essential advantages of adopting the XML
approach. We have discussed in particular the benefits in
creating re-usable namespaced information components or
objects, and how these can be created and validated using
subject-specific ontologies and dictionaries, and enhanced with
appropriate metadata. The role of communities and global
organisations such as IUPAC is seen as crucial in this
endeavour towards creating these key resources. The use of such
XML-based documents opens the prospect of creating avenues for
the reversible flow of data and information between the
scientific publication processes and the discovery, research
and learning processes in molecular sciences, a reversibilty
that has hitherto only been achieved with considerable (and
error-prone) human effort and expense.
12. References
- T. Berners-Lee, M. Fischetti, M, "Weaving the Web: The
Original Design and the Ultimate Destiny of the World-Wide
Web", Orion Business Books, London, 1999. ISBN 0752820907.
For discussion of this in a molecular context, see H. S.
Rzepa and P. Murray-Rust, Learned Publishing, 2001,
14, 177; P. Murray-Rust and H. S. Rzepa, Data Science, 2002, issue 1,
in press.
- D. H. Peapus, H. J. Chiu and N. Campobasso,
Biochemistry, 2001, 40, 10103-10114.
- See http://www.xml.com/pub/rg/117
- See http://www.wr3.org for details of all XML
specifications.
- G. V. Gkoutos, P. Murray-Rust, H. S. Rzepa, and M.
Wright, Internet J. Chemistry, 2001, article 13.
- P. Murray-Rust and H. S. Rzepa, Data Science,
2002, submitted for publication.
- P. Murray-Rust, H. S. Rzepa and M. Wright, New J.
Chem., 2001, 618-634.
- P. Murray-Rust and H. S. Rzepa, J. Chem. Inf. Comp.
Sci., 1999, 39, 928; P. Murray-Rust and H. S. Rzepa,
J. Chem. Inf. Comp. Sci., 2001, 1113; G. Gkoutos, P.
Murray-Rust, H. S. Rzepa and M. Wright, J. Chem. Inf.
Comp. Sci., 2001, 1124.
- See G. P. Moss, http://www.chem.qmw.ac.uk/iupac/ for IUPAC
dictionaries in Web-form. IUPAC home is at http://www.iupac.org/


