CML is a complex language and there are many ways to begin; this is for those who like tutorials. Because CML is flexible, and can cover most apsects of molecular science, it may take a little while to adjust. It will help if you are familiar with the basiscs of HTML and the use of hypertext and linked documents on the WWW.
CML is a formal method of organising information and the result will depend very much on the author's views. It's quite possible for two people to represent the same information in different ways in CML, but it's important to realise that these are often equivalent and can be interconverted by machines or humans. CML can be regarded as a (limited) programming language, (but no more forbidding than HTML!). CML files can be very small (a few lines) or very large - many megabytes with thousands of tags. Indeed, I am planning to write my next book in CML.
Throughout this tutorial we shall show examples of CML files and the way they look in my JUMBO. It's important to realise that CML files can be used without any viewing software, and in some cases without any software at all. Most of the time, however, you will need some software to help create, manipulate, view and transform CML files. JUMBO is able to do most of this, but I hope that much other software will be developed.
The CML examples are denoted by hyperlinks like this [hello.cml] and when you follow the link (click) you will see the raw CML data. (You will find it useful to clone another browser window). For each such file there will also be one or more screen shots from JUMBO. The JUMBO software is written in Java and available as applets on the WWW or as standalone applications. See the FAQ for details. If you are able to run JUMBO, you should be able to reproduce the diagrams (allowing for different windowing systems) and to explore other renderings.
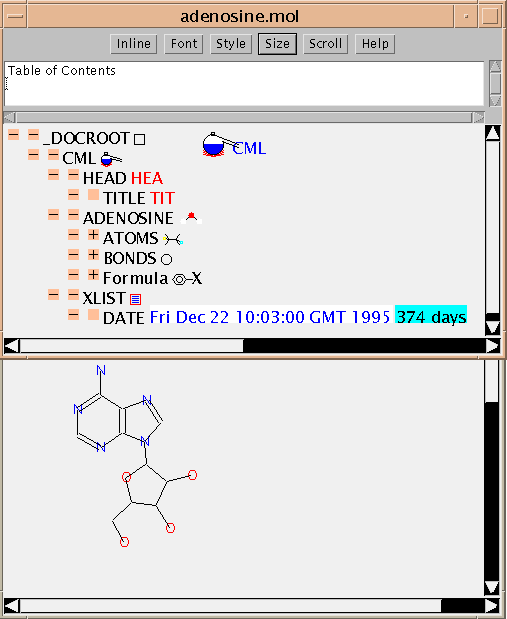
In many applications you won't need to see any CML as it will be 'hidden' within the guts of the application software. So the first example shows the JUMBO reading an MDL Molfile and displaying it. (The command was
java pmr.cml.CMLObj adenosine.moland the result is shown here:

This example also shows that most 'flat files' contain implicit hierarchical information. You may wish to compare the raw file with the picture. (The stereochemistry is not shown - that's a current limitation of the JUMBO depiction, not of CML).
It's straightfoward to produce a CML file at this stage and JUMBO will do this if the _DOCROOT icon square is clicked. Here is the resulting CML file. This contains precisely the same information as the original Molfile and you can see that it's easy to understand, and that the information is much better identified. Don't worry about the details at present; that will be explained later. Note the use of the CONVENTION attribute which allows a future reader (machine or human) to identify what convention is/are used. For example, you would need to refer to the MDL manuals to understand the figures in the BUILTIN="STER" array (although JUMBO will add an implementation in the future). (NOTE: this type of conversion depends on how easily the original file can be interpreted and parsed, not on CML. In some of my converters information has not been preserved because I didn't know what it meant or whether it was significant! It's possible for someone to add this later if required.)
Can a CML file be converted to a Molfile? That depends entirely on what is in the CML file and whether the Molfile can hold that information. For example, objects like sequences, hypertext, diagrams - all of which can be held in CML - cannot be held and so would be lost. All conversion between current file types involves information loss; CML is the only method of ensuring that everything is kept after conversion.
The next two examples show that CML can hold non-molecular information, especially text.
There is some unavoidable terminology here - don't worry about it on first reading
Let's start with the simplest CML file of all: [hello.cml], reproduced here:
<!DOCTYPE CML PUBLIC "-//CML//DTD CML//EN"> <CML TITLE="Hello Benzene"> </CML>In the JUMBO it looks like this:
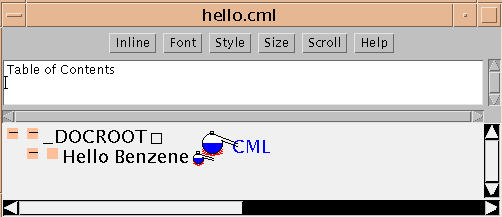
The ELEMENT is defined by the start-tag (<CML>) and end-tag (</CML>). It has no content (in this case), but has a single attribute (TITLE="Hello Benzene"). In general, attributes should describe an element rather than be part of its content, and CML follows this closely. Note that start- and end-tags in CML must always be balanced and must nest neatly. (The only exception is empty tags in HTML such as <BR> or <HR>.)
The JUMBO window renders the information in the CML file precisely. The type of the element is shown by the icon (a small alembic for CML elements). (The large alembic is the CML logo.) JUMBO labels elements with their TITLE attribute if they have one, but otherwise with the type of element (technically known as generic identifier or GI).
Some final technical points:
This is a larger example and shows the power of the TOC and the mixing of different sorts of information. Here is the hello1.cml source file and here is the screen shot of what it produces:
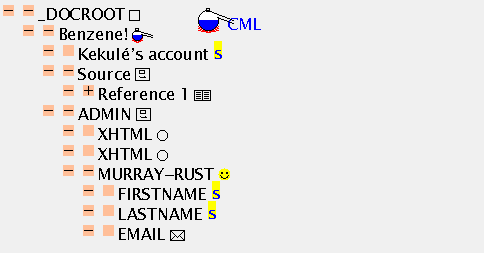
At this stage it's worth comparing the TOC with the hello1.cml source file. You will note that JUMBO has (deliberately) hidden quite a lot of the material so as not to confuse you. An inevitable consequence of marking up your information is that there is suddenly much greater apparent complexity, and friendly navigational tools will become crucial. It's also important for authors to think more carefully how they present information.
Why go to the complexity of 10 lines (<BIB> ... </BIB>) when
it's shorter to write and read:
James Kendall, Great Discoveries by Young Chemists, p94 (1953) publ:
Thomas Nelson and Sons Ltd?
In practice it can be very difficult to
parse such text (what are the volume numbers, pages, years, etc.?) and
it's almost impossible to search it. In CML the containment and
markup make it easy to search for, say, a publisher called
Nelson rather than an author. It's still possible to display
the information succinctly and we'll see that in later examples.
The next figure
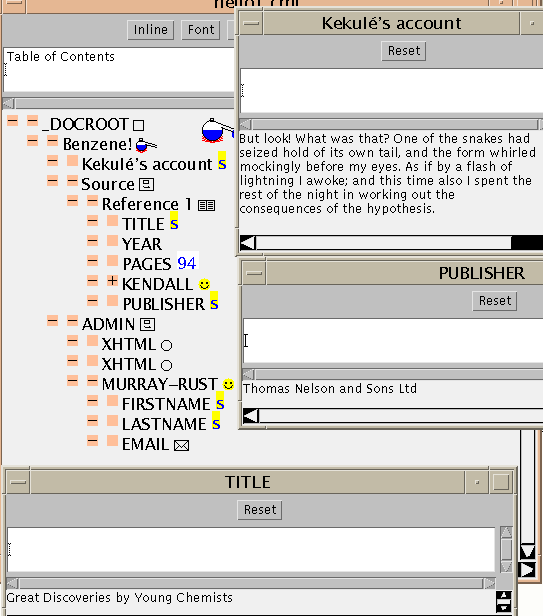
A feature you may have noticed is the accented e (é). This is provided by an SGML entity with the symbolic name eacute. When the application parses the file it looks for strings of the form &something; and, if possible, replaces them by their value. In this case, eacute is defined in ISO-Latin 1, and is also implemented in the Java character see and all commonly used browsers. There are lots of other entity sets (many from ISO) for Greek, maths, etc., but as yet relatively few browsers or fonts have glyphs for them. Happily there is now much emphasis on extended character sets (e.g. ISO10646) and SGML has been designed to support them.
We shall now explore in detail a simple molecule, and then its mass spectrum. The molecule is held in the CML file mol.cml, which is displayed as a TOC:
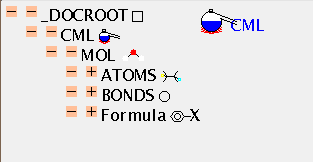
There is no mandatory field in ATOMS, but ELSYM will almost always be present. This is an ARRAY of symbols (in this case 11), separated by whitespace. Normally, as here, they will be from the periodic table but others are permitted (such as dummy) and hopefully an agreed list will evolve. The list is terminated by the closing tag and the count can be deduced by the application (there is a (redundant) SIZE attribute if required).
The ATOMS have X2 and Y2 ARRAYs to provide drawing coordinates. (There are separate attributes X3, Y3 and Z3 for 3-dimensional coordinates which can coexists with 2-D coordinates if appropriate). There is also a list of the atomic charges (all zero in this case). Note that all ARRAYs within ATOMS have to be the same length, or the file is invalid. There are about 30 possible values for the BUILTIN attribute, and it will be expected that application software should be able to deal with all of these in some way (e.g. by drawing, producing tables, etc.). There is a simple, robust, mechanism for adding additional atomic properties as ARRAYs when necessary.
BONDS follow a similar format. The normal way to specify a bond is to identify the atoms at the ends (ATID1 and ATID2). These may be serial numbers (as here) and always start from 1 (not zero), but may also be any unique identifer (e.g. 'C13A'). The bond ORDER may have various conventions and if it is other than 1, 2, or 3 this should be specified (e.g. CSD aromatic is -5).
The formula of a molecule cannot always be calculated from its coordinates (there could be missing atoms, for example), and FORMULA gives a variety of tools. The most common are 2-d connexion tables, and SMILES. Other conventions will need to be described with care.
The TOC is shown in more depth here:

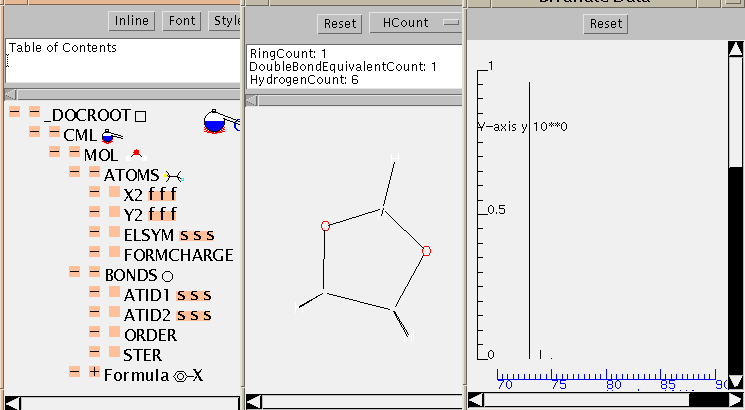
This application contains tables of isotopes so it is also possible to calculate the isotopic variation of the molecular mass. This is shown as a simulated mass spectrum (with hydrogen loss) and can be compared with the experimental (see below).
You might want to reveist the first example and see how the CML files has been constructed.
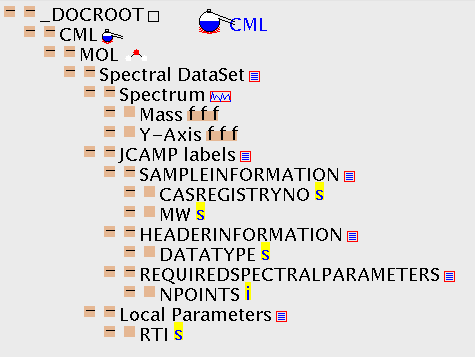
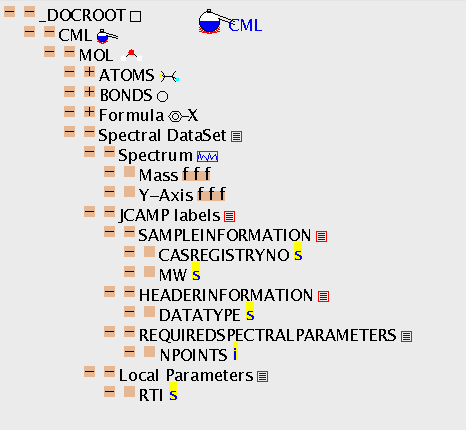
CML has been specifically designed to cater for experimental data and we show a mass spectrum as an example. It was obtained in the JCAMP standard and automatically translated to CML in JUMBO to give ms.cml. (I have pretty-printed this for visual impact, but this doesn't affect the content.) This introduces a key feature of CML: extensibility through external dictionaries or glossaries. Thus XVALUES is defined in the printed JCAMP standard so its use is precisely defined. An application program which can display spectra from CML files could be expected to understand the role of all these terms.
The spectrum itself is contained in an XLIST with BUILTIN="SPECTRUM", and the attributes are doing the job of definining thr information. (You could argue that SPECTRUM is sufficiently important to have its own tag and it would be reasonable for the spectroscopy community to develop its own CML-compatible DTD in the future). At present the generality of CML is kept by not multiplying tags, but putting a high emphasis on the BUILTIN attribute values. Notice the use of FLOAT to define real numbers.
The spectrum has two more attributes: CONTENT=GRAPH declares that the content of XLIST (which can be very flexible) is constrained to be representable as a graph (i.e. have two ARRAYs of equal lengths). (It's not really necessary as BUILTIN="SPECTRUM" implies that anyway). DISPLAY="BAR" is a hint to use a bargraph display rather than a continuous line - this is advisory only as it doesn't form part of the content).
Note how numeric data can be mixed with other information (although the spectrometer hasn't output very meaningful values!). CML can faithfully capture this and use it in other applications.
Here is the TOC:

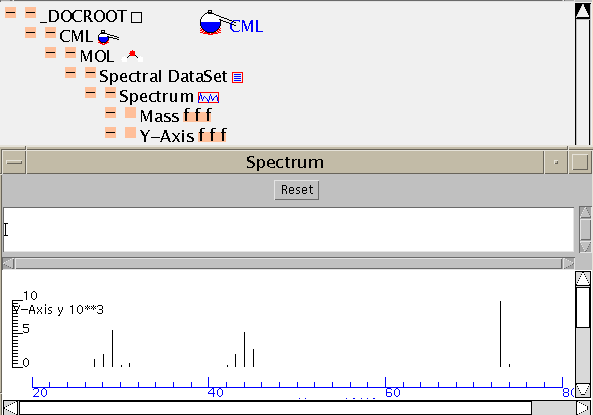
CML has very few limitations on the information structure and so it's easy to combine simple components into larger compound documents. In compnd.cml the molecule and its spectrum have been combined by simple manual cut-and-paste. (Shortly this will be possible with graphical tools). The resultant TOC shows the simple combination:
This file you are reading is CML! - because CML includes a subset of HTML2.0. Assuming that it's XML-like (i.e. tags are balanced, attribute values quoted) it's possible to display it in JUMBO. Here's a shorter file faq.html (an early FAQ) and here's its TOC:



Although simple, these examples have shown the variety of documents that can be constructed in CML. After mastering them (especially if you have JUMBO) it's probably best to browse the wider and more complex range of examples
© Peter Murray-Rust, 1996, 1997