At present I have only processed FASTA and SwissProt
files into CML format, but
it is easy to do the same for other formats
CML has a SEQUENCE ELEMENT designed specifically for macromolecular
sequences. It has flexible content but should be straightforward for
linear polymers such as proteins and nucleic acids. There is, as yet, no
simple convention for modified macromolecules (e.g. glycosylated proteins)
or for branched and cyclic polymers (e.g. oligosaccharidea). However,
since CML is capable of holding many MOL elements in a document, including
hierarchical containment it should be possible to develop powerful systems
for referencing different types of component in oligomers.
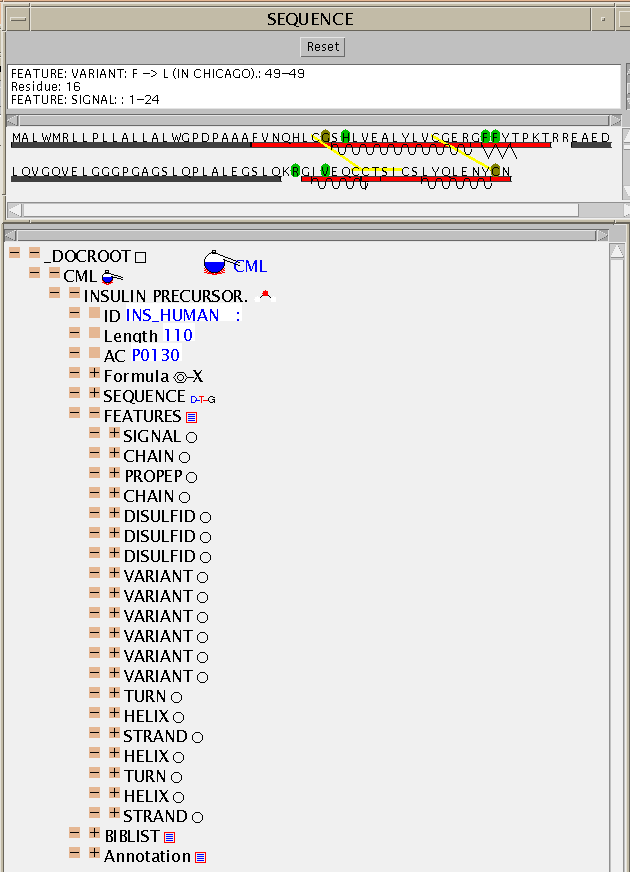
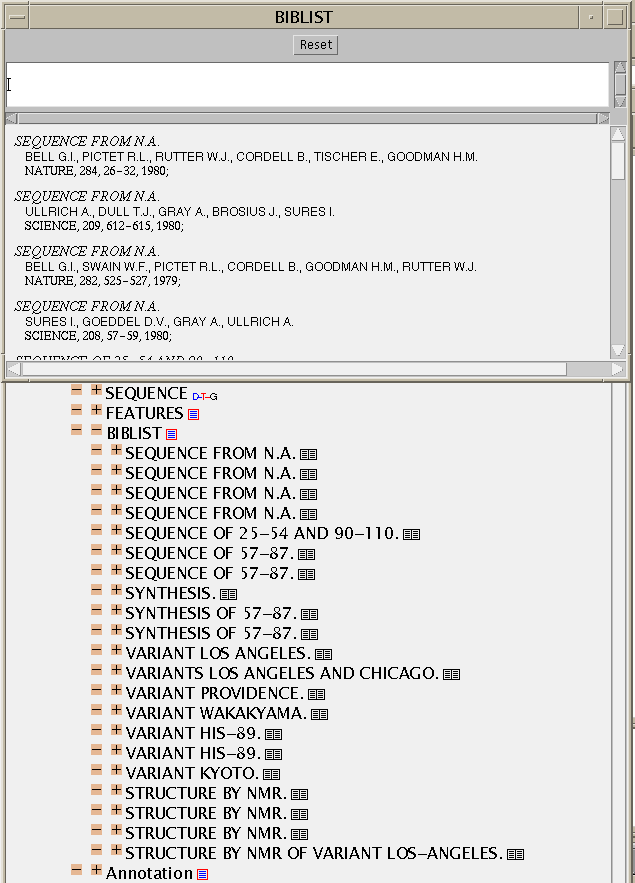
An important aspect of macromolecules is the FEATURE, for which a special ELEMENT is also provided. Normally FEATURE is an annotation in that it describes the relevance of part of the sequence, but sometimes it also indicates molecular connectivity (e.g. disulphides, glycosylation, or metal binding). It is too early to suggest all the ways that FEATURE can be used, but it will often make use of the ability to address parts or an object ('Sub-addressing'). This is a generic feature in CML and allows an application programmer to let one object (or parts of it) interact with parts of another. The FEATURES, therefore, can sub-address the SEQUENCE if required, though the action and interpretation is normally left to the application.
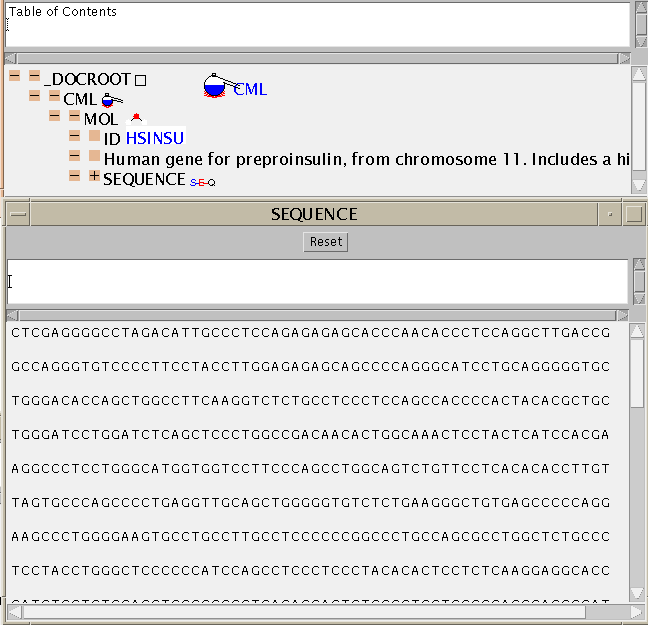
FASTA is a simple format for holding sequences, and the example here is from the EMBL distribution (hsinsu.fasta). Essentially it only contains an ID, a description and a sequence, so the CML TOC is very simple:

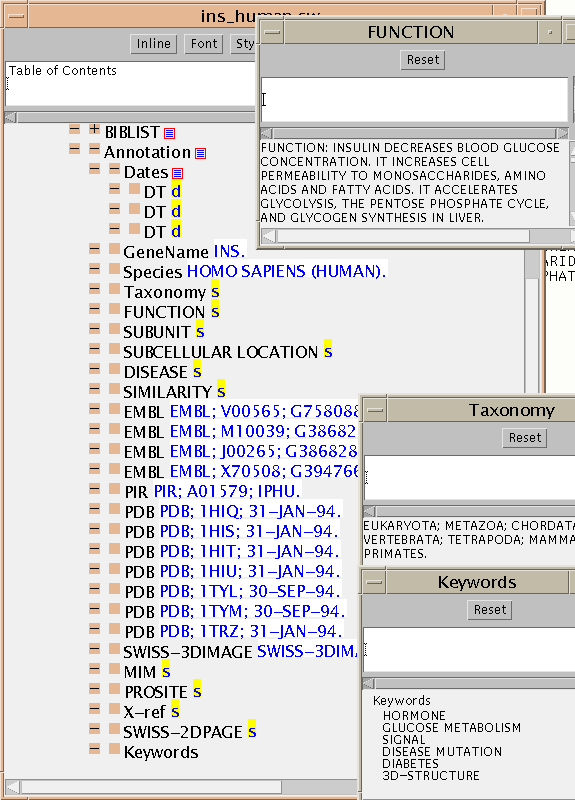
CML has a SwissProt parser which understands 'most' of the records in the file. Particular emphasis has been put on showing the use of FEATURE, BIBliograpghy and, sequence, but many of the other keywords have been objectified (the main omission being the administrivia).
This is the SwissProt entry for human insulin and its automatic conversion to CML ins_human.cml. The TOC
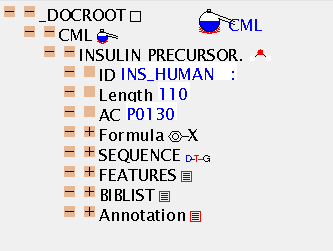
CML pays great attention to bibliography and citations and JUMBO can
render everything in the SwissProt file.
Finally we show some of the annotations (a mixture of keywords, comments,
dates and other administrivia). Note how the simple scalars (e.g.
GeneName, Species) can be toggled on or off inline. Those with larger
content (e.g. Comments such as FUNCTION) are displayed as a text box.
In some cases the information is a list of items (strings, numbers, etc.)
as in the KEYWORDS.