Protein Structure
CML is particularly; useful in managing protein structures as there
is a complex mixture of information in most 'flat' files such as
those from the Protein Data Bank (PDB). These include administrivia,
citations, annotation, sequence, crystallography and 'small molecules'
as well as the 3-D coordinates of the protein itself. Unfortunately
most of this information is very rarely used because there is no
useful way of managing it. (How many molecular modelling packages
manage citations satisfactorily?).
This example includes two protein structures from PDB, each with
points of interest for the markup. Remember that JUMBO is not
intended to be a full molecular viewing and manipulation program, so
don't expect high quality rendering! Note that the CML parser
will only fully parse strict PDB files (there are a large number of mutant
'PDB-like' files in which the only communality is that they contain
'ATOM' cards; JUMBO does what it can with these).
Insulin
The PDB file
1insmini.pdb is a cut-down version (only a monomer
is selected). The TOC
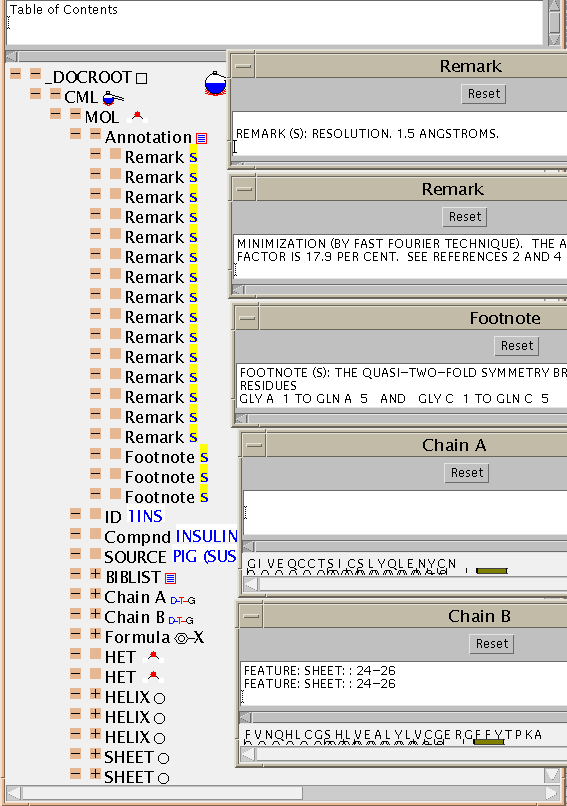
shows the large variety of information collected and standardised for
a typical PDB entry. If you are familiar with PDB files, you'll see the order
is preserved as much as possible.
- ID is extracted from the HEADER
- COMPND and SOURCE are preserved as discrete objects. Wherever possible
CML encapsulates existing pseudo-objects in an XVAR, ARRAY or XLIST. It is
often possible to type these as (say) FLOAT or DATE. The semantics are
added through DICTNAME and CONVENTION records so that CML-compliant software can
apply appropriate methods to the objects. Thus CONVENTION="PDB" refers users
to the PDB user manual and DICTNAME indicates the precise keyword in there.
We strongly recommend this approach for anyone using CML documents with
objects outside the range of this documentation.
- The PDB uses REMARK for annotations and comments. This is presently formatted
à la FORTRAN but would benefit immensely from additional markup
(e.g. for residues, tables, etc.).
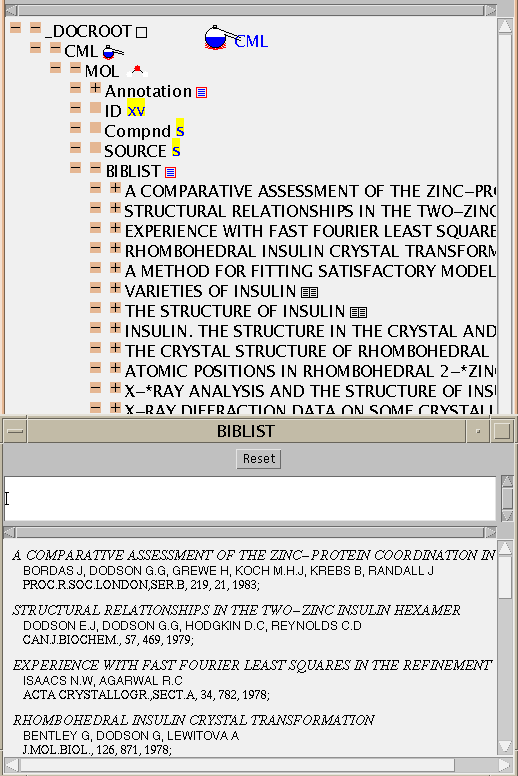
- The PDB also uses REMARK for citations, and this is a tedious parsing problem.
The CML parser has done this, so that all citations are fully structured BIB
objects. under BIBLIST
- The sequences under SEQRES have been transformed to SEQUENCE objects, and
labelled with their chain IDs ("Chain A"). Compare the displays with the
SwissProt entry (remember that that was the
preproprotein and that the B chain comes before the A!).
- The HET object refers to a dictionary of molecules and until this is
translated into CML , HET is retained as a simple text string
.
- The secondary structure features (HELIX, SHEEET, TURN) are captured as
FEATURES. (Maybe they should go under an XLIST container?). They contain
pointers to other objects and subobjects in the CML document and the
addressing is being developed
- SSBOND is captured as a FEATURE (DISULFID) and can be used to subaddress
the appropriate atoms and residues. (This and subsequent objects are displayed
in a later figure)
- SITE is also captured as a FEATURE and can use ARRAYs of (sub)addresses to
identify all the components
- CRYST incorporates the CRYST, SCALE and MTRIX records. CML uses ARRAY and/or
XLIST to manage matrices
Here is an expanded version of the citations (BIBLIST)
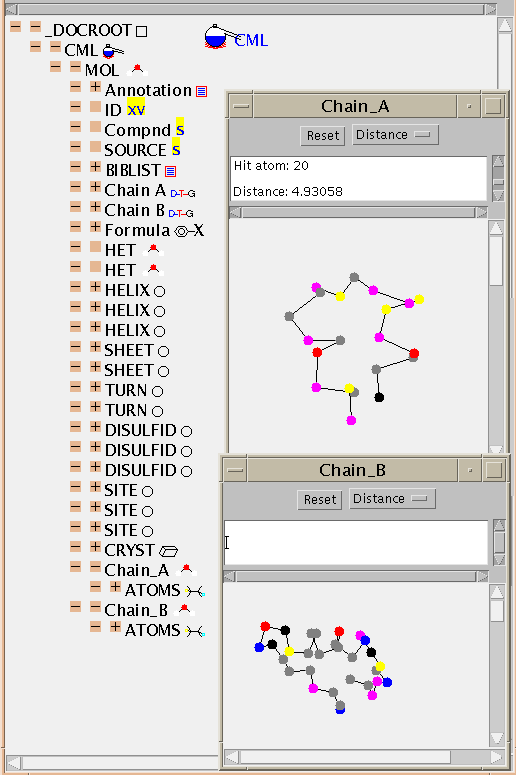
CML allows MOLs to contain other MOLs which is valuable for macromolecular
structures. In this case the two chains of the molecule are held separately
and it's up to the application how they are treated (for this example JUMBO
is displaying the chains in separate windows with different orientations and
scales, but they could be combined and ganged together).
The resulting CML file is
1ins.cml. It's no larger than the original PDB file
and reads in much quicker as it doesn't need parsing.
Flavodoxin
This is an example of a protein with a single chain, but a small molecule
ligand. The (edited) PDB file is
4fxnmini.pdb and a typical screenshot is
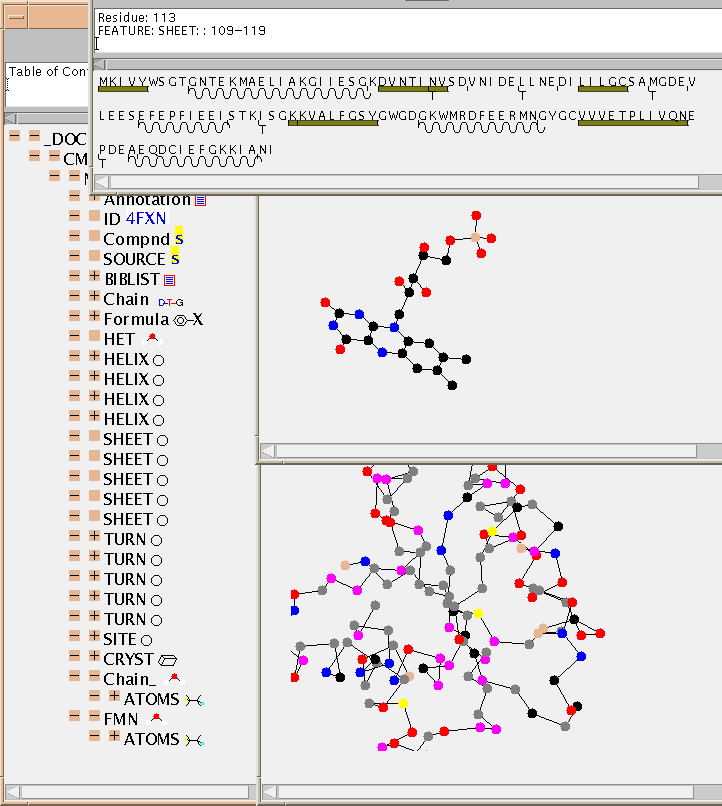
where you can see the ligand (FMN - middle right), and the annotated
SEQUENCE ( top window - sine curve = HELIX,
bar = SHEET). The reader has just clicked on the bar under "VVVET...".
Back to index
©
Peter Murray-Rust, 1996, 1997




