Glossaries are very important in CML as a way of resolving the semantics of objects, such as XVAR. Ideally every marked-up object should either be hardcoded into CML or point to an entry in a glossary. As CML develops these glossaries can contain not only textual descriptions, but also data (e.g. molecular coordinates or physical quantities) and methods. The first steps in this process are the use of scientific Units in a glossary.
Several molecular glossaries have been or are being marked up and the latest position can be found in the Virtual Hyperglossary Project. Here we shall give a small number of simple examples.
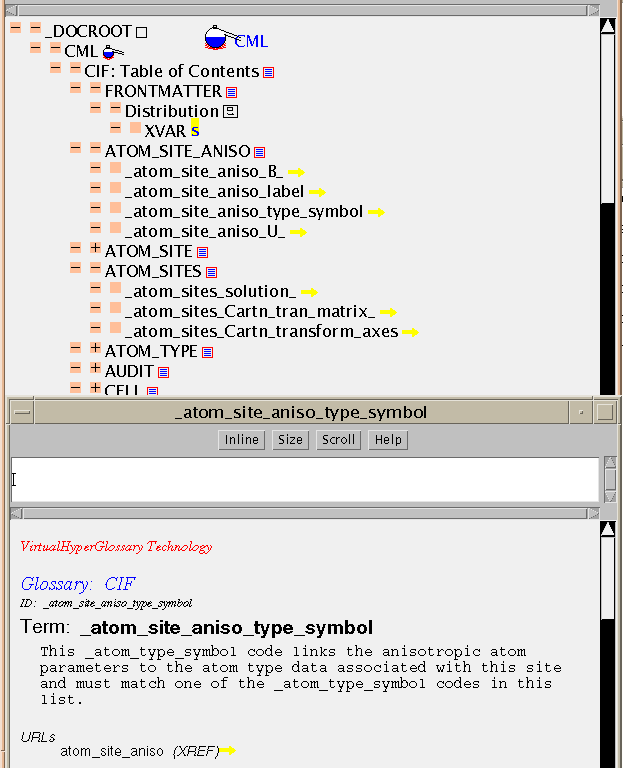
The International Union of Crystallography (IUCr) has sponsored the development of a set of dictionaries (a combination of glossaries and data dictionaries). Here is a demo of the Core dictionary (1993 version). The glossary consists of about 240 terms organised into categories, and a short Java/CML program has parsed this into a table of contents (ciftoc.cml). The main body of this is a list of terms arranged under categories (XLIST is used to contain each group). However the terms are not part of the document, but are separate as in a typical entry: ( atom_site_aniso_type_symbol.cml). Looking at this you will see that its components are XVARs, with an XHTML for the definition, and some ADMIN nodes. The key components are the ID and TERM (held in XVARs) and the definition (held in XHTML). The owner of the entry ("sourceIdentifier") is held in an ADMIN node.
The TOC contains a list of pointers to these entries, of the form:
<XVAR TYPE="URL" MIME="chemical/x-cml" TITLE="someTitle">someFilename</XVAR>In CML TYPE="URL" implies that the XVAR contains pointer information as its content, and that the TYPE of the information is given by the MIME attribute (i.e. the target is a CML file). (The target can be any file type, but the application program may not always be able to process it! This is a much safer method than relying on suffixes).
This is resolved by JUMBO in the TOC where some of the terms have been
unhidden.

Since the target of a CML/URL pointer can be any type of file, molecular
information can be contained in glossaries. In the Principles of Protein
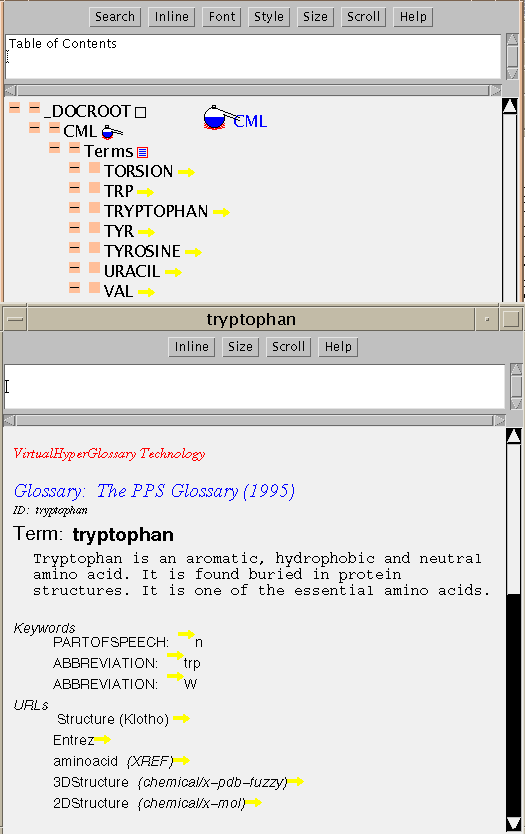
Structure virtual course, we developed a hyperglossary for protein
structure terms and I have now converted this to CML. Part of the TOC is
shown with the entry for tryptophan
(tryptophan.ent):
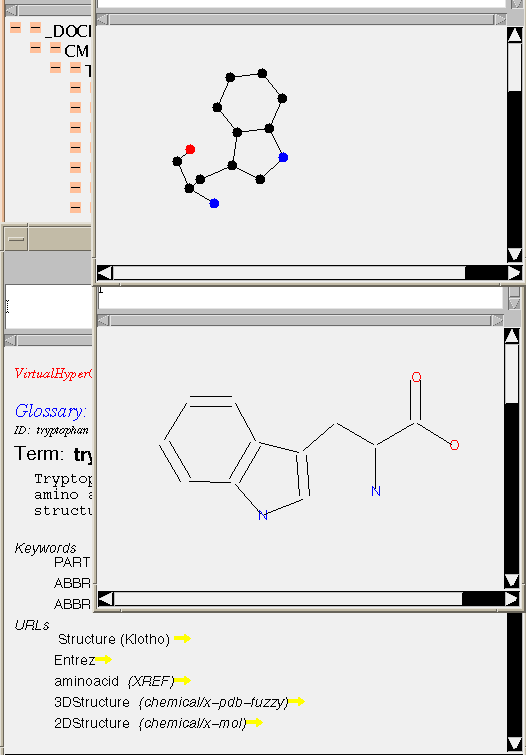
By adding TYPEs and other constraints to hyperlinks, CML makes it possible to represent complex information architectures, and to make it easier to develop and test the software required.