Instructions for Chemical Information Technology
These pages represent detailed instructions for the techniques described. Some
techniques are deemed sufficiently "intuitive" that no details are given here.
In other cases, the information provided by the supplier itself is sufficient,
and is not replicated here. To access each individual information point, click on
the icon you see. To find out more about the information provider, click on the blue
hyperlink next to the icon. The strip  means you can return to
an overview of the "Information booth".
means you can return to
an overview of the "Information booth".



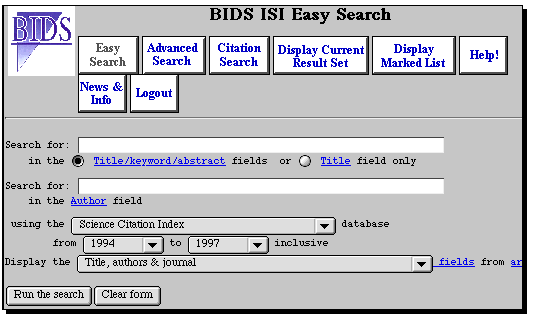
BIDS
This service is available via either a Web page or via a terminal emulation program called Telnet.
The screen-dumps shown here are for the Web interface. To see those for the
Telnet interface, go here.
Login with
account iic02p (the password is available from members of staff) and select the ISI service.
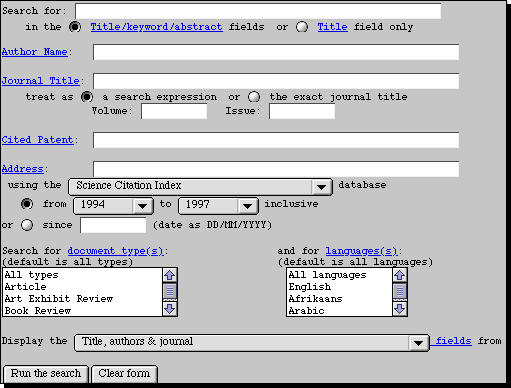
More detailed instructions are summarised in a small manual
available from the
chemistry library or the on-line office in the Lyon Playfair laboratory.


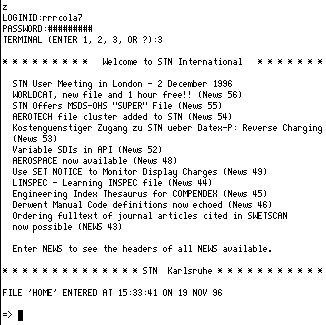
CAS On-line
This will enter a Telnet session. Type z as the first thing you enter, followed by
an account number and password (this is available on request) followed by 3 for the
terminal type.
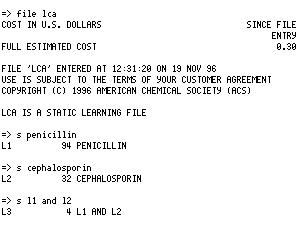
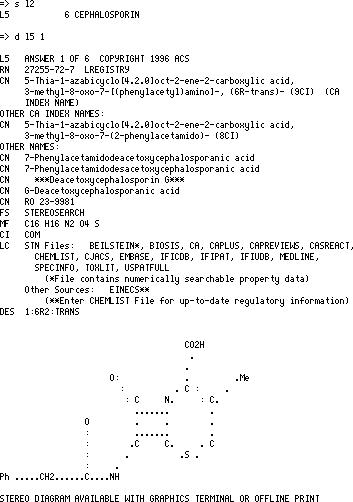
You will next have to specify the database file you want by type FILE LCA (the Learning
CAS bibliographic file)
or FILE LREG (the Learning Registry file of substances)
To use the CAS system enter the following series of commands;



Libertas
Again the Telnet program is used to access the College catalogues;





 This information provider was one of the first
companies to offer a "Web" interface to their databases. Here you can see it
in action on "sampler" datasets. A number of interfaces are offered in this
service, including the World Drug Index, and the "Savant" system.
This information provider was one of the first
companies to offer a "Web" interface to their databases. Here you can see it
in action on "sampler" datasets. A number of interfaces are offered in this
service, including the World Drug Index, and the "Savant" system.
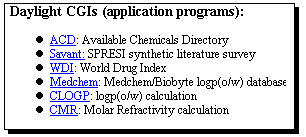
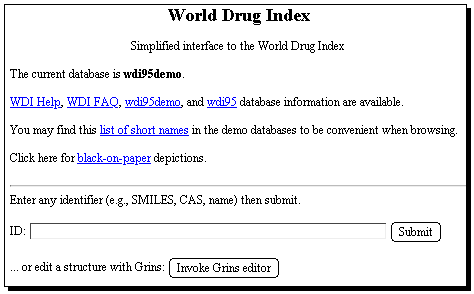
Start with the WDI database, entering a suitable search term;
The results of a keyword search are displayed on the screen.
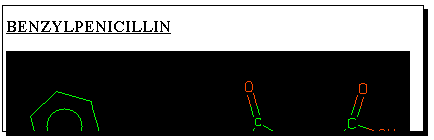
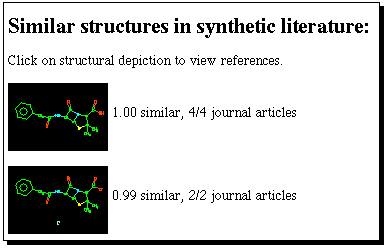
Clicking
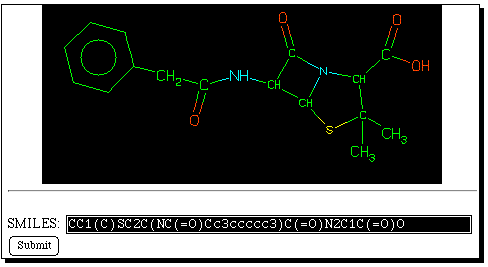
on the "thumbnail" graphic of any molecule found will reveal
further properties, one of which is the so-called SMILES
string. This is a powerful and popular method of representing
molecular structure as a sequence of simple characters. It
is also one of the few methods for transferring molecular
structure definitions between different programs. To illustrate
how this works, select the SMILES string shown below the
structure, and "copy" it to the clipboard using the "edit" menu.
Now select the "back" button on the WWW browser, and enter
instead the "Savant" database. The query SMILES string
can be pasted into the keyword search field. Savant produces
a "similarity" search which finds bibliographic references
to recent literature relating to the synthesis of compounds
either identical to, or similar to the SMILES search query.
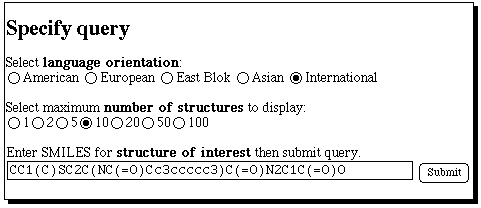 Select a compound which may look interesting, and
investigate one or two recent literature references to
its synthesis;
Select a compound which may look interesting, and
investigate one or two recent literature references to
its synthesis;

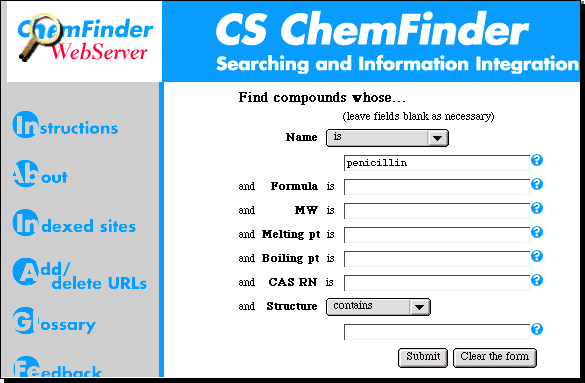
 This information provider was also one of the first
companies to offer a "Web" interface to their databases. See what they have to say
about penicillin.
This information provider was also one of the first
companies to offer a "Web" interface to their databases. See what they have to say
about penicillin.

 When Beilstein Commander starts up, you will be confronted by a login-prompt. You will
be issued with account numbers and passwords during the course.
When Beilstein Commander starts up, you will be confronted by a login-prompt. You will
be issued with account numbers and passwords during the course.
 Due to a programming error in this program, the molecule window is empty.
Click on
Due to a programming error in this program, the molecule window is empty.
Click on  a second
time, and this will be replaced with a molecule query.
a second
time, and this will be replaced with a molecule query.
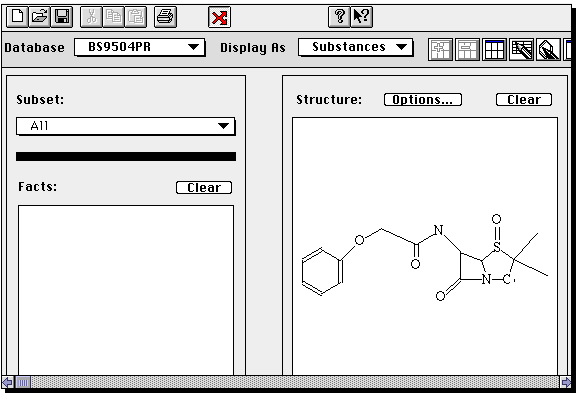 If you wish to edit or modify this structure, double click on the molecule, whereupon
a structure editor window will open;
If you wish to edit or modify this structure, double click on the molecule, whereupon
a structure editor window will open;
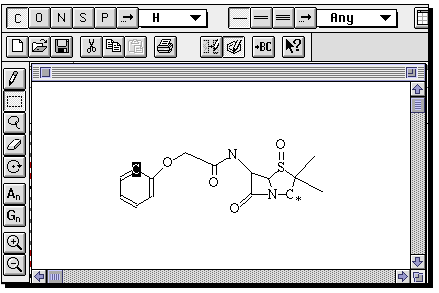 The important thing about this structure, is that missing valencies are assumed
to be hydrogen rather than generic substituents. To allow a search to proceed
on the assumption that any substituent can occupy a free site, go to the capture tool
(the dotted square icon), then highlight an atom (or several, by dragging the capture
box) and from the Query menu, select "Free Sites". This places a star against the atom(s).
If you do not do this, you may not find any structures. Other editing operations are
(sort of) intuitive, and it is suggested you explore them. Once happy with the structure,
click on the "BC" button along the top to return to Commander.
The important thing about this structure, is that missing valencies are assumed
to be hydrogen rather than generic substituents. To allow a search to proceed
on the assumption that any substituent can occupy a free site, go to the capture tool
(the dotted square icon), then highlight an atom (or several, by dragging the capture
box) and from the Query menu, select "Free Sites". This places a star against the atom(s).
If you do not do this, you may not find any structures. Other editing operations are
(sort of) intuitive, and it is suggested you explore them. Once happy with the structure,
click on the "BC" button along the top to return to Commander.
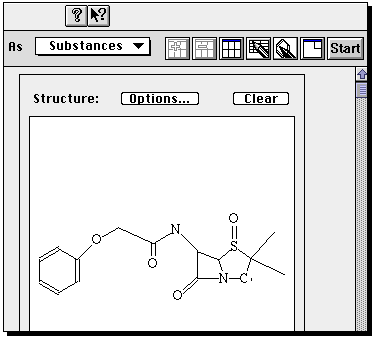 Now click on Start to
commence the search.
You will be told how many hits are found (for an unmodified structure
query it should be between 100-200!). A new program called Display Hits starts up
(by this time, your Macintosh might be suffering from a lack of memory, and you
may need to shut down any non-essential programs). From menu View, select Short display to
get a preview of structures found.
Now click on Start to
commence the search.
You will be told how many hits are found (for an unmodified structure
query it should be between 100-200!). A new program called Display Hits starts up
(by this time, your Macintosh might be suffering from a lack of memory, and you
may need to shut down any non-essential programs). From menu View, select Short display to
get a preview of structures found.
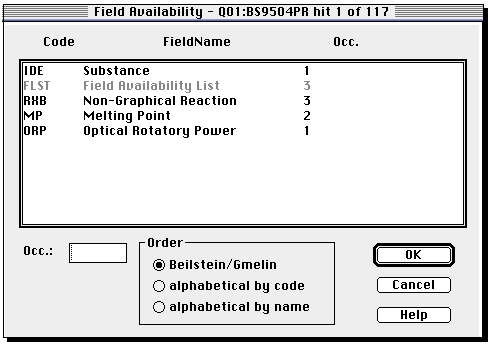
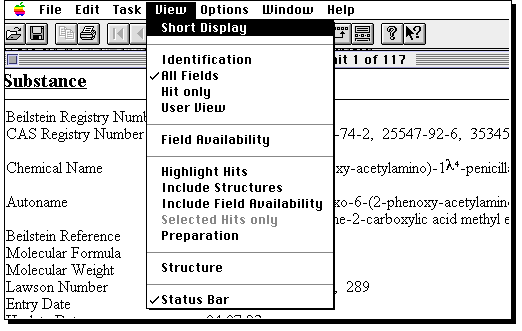 You can select individual entries by clicking on them.
Go back to full display to obtain data such as optical rotation or melting point.
You can select individual entries by clicking on them.
Go back to full display to obtain data such as optical rotation or melting point.

 Clicking on the ISIS logo activates a program
called ISIS/Draw. This should already contain a reaction query defined
for you.
Clicking on the ISIS logo activates a program
called ISIS/Draw. This should already contain a reaction query defined
for you.
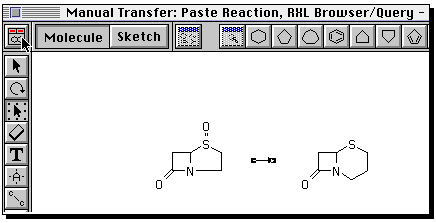
You can if you wish define an entirely new reaction. To do so,
proceed as follows. Build the reactant using ring templates,
and suitable bond tools. To insert heteroatoms, select
the "A" tool, click on the atom, when a small box should appear,
and type the atom symbol from the keyboard.
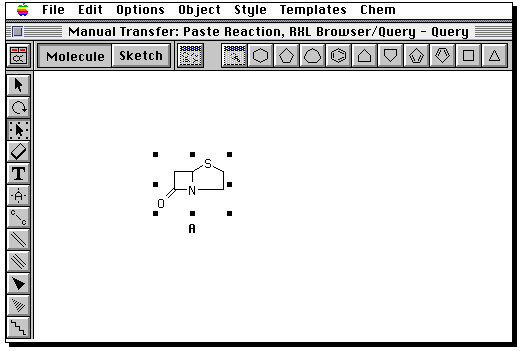
Draw the product in the same manner. Then go to the "box" tool,
and "select all" from the edit menu. Now from the "Chem" menu,
select "reaction".
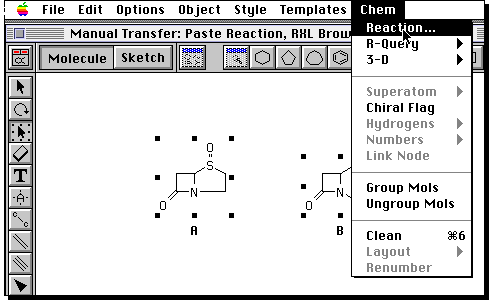
You now have to define the relationship of the two molecules
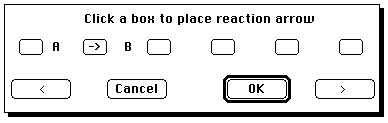
A reaction is defined. Now you have to launch ISIS/Base. Because this
requires a lot of memory, you will have to close any other applications
that might be running (including Netscape!). Launch the program from the
Apple menu in the "chemistry programs" sub-menu. Once open, select
"database" from the "file" menu. Open RXN browser. You will need to
provide a user number and password. Ask a member of staff for this.
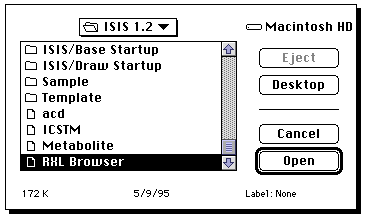
Once Base is up and running (it may take about 2-3 minutes to achieve this
state) return to the Draw program. Select ALL and then COPY from the Edit menu,
return to Base, and PASTE the diagram in.
The reaction should now appear in the Base window;
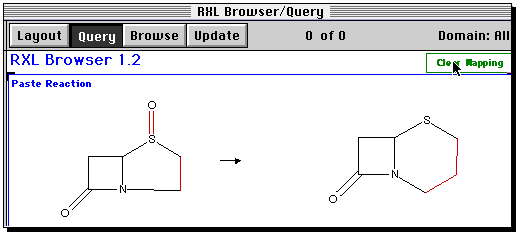
Some bonds will be highlighted to indicate a possible
bond mapping between the two structures. This guess is likely
to be wrong, so the safest course of action is to clear mapping.
Now from the Search menu, select RSS (reaction sub-structure);
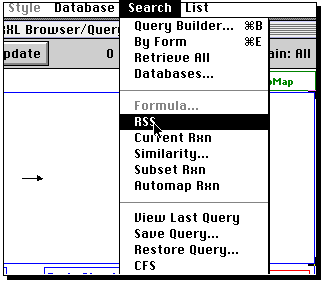
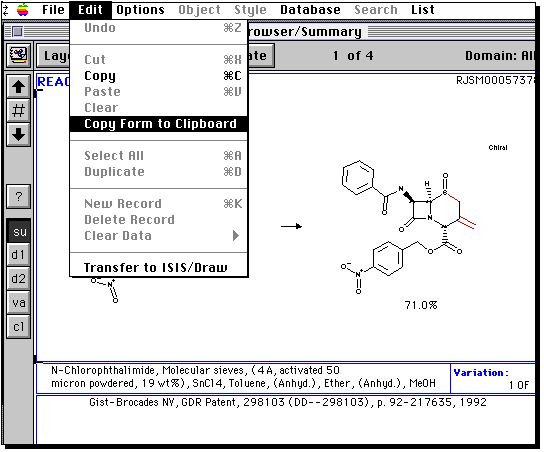
A list of "hits" will appear. Select the "su" button, and if you wish
scroll down the list of hits with the "down" button. If you wish to
copy an entire hit, select the "copy form" item from the edit menu,
and then paste this into a word processor. (since you may have stopped
all other programs due to memory, you may now have to start Claris up again).

 You will need to search the Cambridge
crystal structure database for the penicillin and cephalosporin sub-structures.
A penicillin search query has been pre-prepared for you. To invoke it,
You will need to search the Cambridge
crystal structure database for the penicillin and cephalosporin sub-structures.
A penicillin search query has been pre-prepared for you. To invoke it,
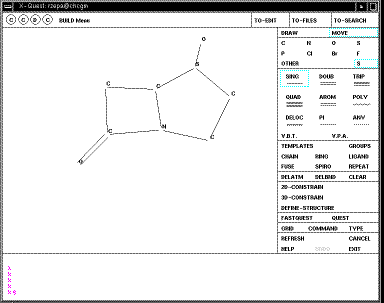
If you are drawing a molecule from scratch, go to the BUILD menu.
You will notice that there is no common standard for
drawing molecules on screen (unfortunately). With Quest, a click
on the screen draws the atom selected (by default C,
but in the example below
currently S). Further clicks add a further atom to the last atom. If you
want the next atom not to be added to the previous, select MOVE first. To
convert a single to a double bond, select DOUB, then MOVE and click on the bond
desired. To convert a carbon to another atom on the menu, select the type from
the screen menu and then MOVE before clicking at the desired atom. If the atom
is not present on the menu, click on OTHER. Errors can be corrected using
DELATOM or DELBND, or in extremis CLEAR which removes the entire molecule.

When you are happy with your structure, click on DEFINE STRUCTURE and then
QUEST;
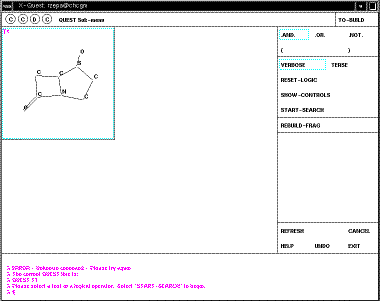
Your defined search operators are shown as T1 (T2 etc). In this case they are
sub-structures, but many other definitions are possible, including author
names, formulae, etc. It is possible to combine several of these using logical
operators. In this case none of this is necessary. Just click on the T1 box to
select this item, then START-SEARCH. As soon as a "hit" is found, it is
displayed on the screen;
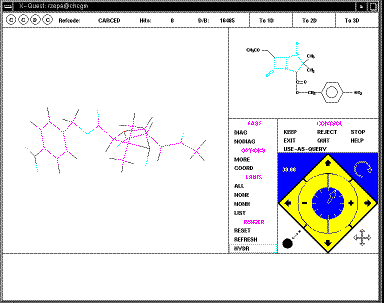
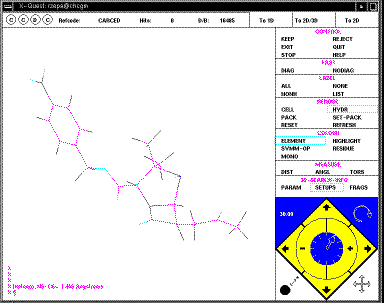
The structure can be rotated using the four small arrows in the bottom right
corner of the menu (you might wish to discuss in your writeup the pros and cons
of this method compared with those found in other programs; again no common
standard applies).
At this point, you have to either KEEP or REJECT the
structure before the next one is displayed. If you KEEP the structure, its
co-ordinates are written to a file on disk (called penicillin.dat in
this case). Continue to the end, when the exit from the database will occur. To
perform a further search, type Quest <name of search> from the console
window of the workstation, as shown in an earlier diagram above. At any stage
you can also go to the alternative 3D projection, in which bond lengths, angles
etc can be displayed on the screen.

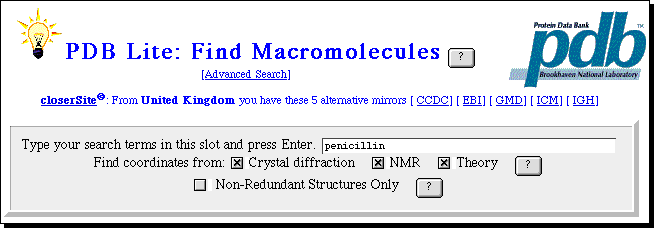
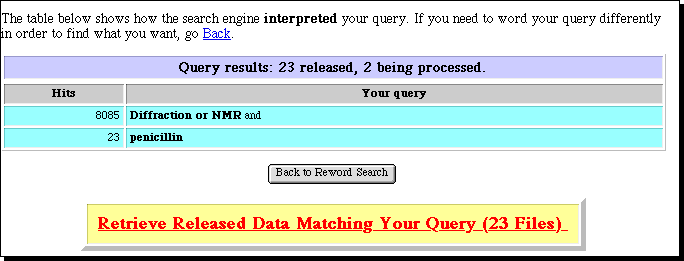
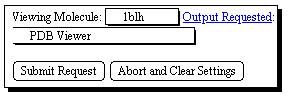
 This enables a simple keyword search of the PDB archives, with the result
displayed in a window on your screen. Be aware that the PDB files can be quite
large, and that response times for their retrieval are likely to be better in
the morning than the afternoon. A mor sophisticated interfaces is avalailable
via the European Bioinformatics Institute.
This enables a simple keyword search of the PDB archives, with the result
displayed in a window on your screen. Be aware that the PDB files can be quite
large, and that response times for their retrieval are likely to be better in
the morning than the afternoon. A mor sophisticated interfaces is avalailable
via the European Bioinformatics Institute.

 This represents a virtual library of structural and chemical information, presented
in the style of a textbook of "knowledge". Can you find any entry for penicillin?
Try also browsing throught another virtual laboratory under construction; VChemLab
and in our NMR Tutorek.
This represents a virtual library of structural and chemical information, presented
in the style of a textbook of "knowledge". Can you find any entry for penicillin?
Try also browsing throught another virtual laboratory under construction; VChemLab
and in our NMR Tutorek.


Electronic Conferences
Unlike real conferences, electronic versions can be easily
indexed. The three ECTOC conferences,
hosted by Imperial College during
1996-1997 were the first such conferences to support
index searching. Find out if penicillin was mentioned in any of them.

 Browse through the Chemical Communications
Browse through the Chemical Communications


 The pasword for entering the course discussion area is available from
demonstrators. Enter this password, and then register yourself to the forum.
Please behave responsibly in this area, since what you do can be seen by
others.
The pasword for entering the course discussion area is available from
demonstrators. Enter this password, and then register yourself to the forum.
Please behave responsibly in this area, since what you do can be seen by
others.


![[BACK]](BackBtn.gif) ------------- Page 7 of 9 -------------
------------- Page 7 of 9 -------------
![[NEXT]](NextBtn.gif)
©H. S. Rzepa and ICSTM Chemistry Department, 1994, 1995, 1996, 1997.

 means you can return to
an overview of the "Information booth".
means you can return to
an overview of the "Information booth".
































 a second
time, and this will be replaced with a molecule query.
a second
time, and this will be replaced with a molecule query.





































![[BACK]](BackBtn.gif) ------------- Page 7 of 9 -------------
------------- Page 7 of 9 -------------
![[NEXT]](NextBtn.gif)