  
Screening for Activity
On the assumption that the library will hold some sequences that
will be tailored for activity against particular proteases of interest, the task
is to find which library component has activity. Activity is characterised by
tight non-covalent binding, and so the enzyme itself can be used as a probe to
discover which members of the library show activity. It is possible to tag the
protease enzyme in such a way that the presence of this protein is characterised
by a colour reaction. Addition of the protein to a suspension of the library
will then cause any resin beads that bind the protein to be coloured dark blue.
These beads can be easily visualised under a low power microscope, and separated
from the non-binding sequences.
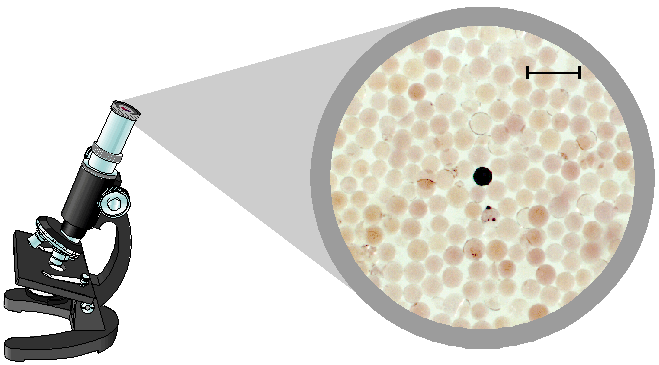
Under the microscope, beads holding sequences that bind tightly
to the protease enzyme become highly stained. In the actual micrograph shown
above there is a single active bead (center) amongst a large number of inactive
beads. The scale bar is 0.2 mm. Approximately 1 in 1000 of the library members
were found to show activity, illustrating the selectivity of the interaction and
the power of library techniques to identify active components.
| 