Introduction
Understanding the extensive network of complex in vivo protein-protein interactions executing the cell cycle and controlling cell structure and function is a crucial aspect of cancer research. In collaboration with the Protein Folding and Assembly Team under Professor Keith Willison at the Institute of Cancer Research we aim to further knowledge in this area, utilising the physical chemistry and optical spectroscopy capabilities of the Molecular Dynamics Group in this highly relevant biological context.
Our work in this regard is focused on the eukaryotic chaperone CCT (Chaperonin Containing TCP-1). CCT is a chaperone positioned at the heart of this biological area, folding the primary cytoskeletal components actin and tubulin and regulating the activity of major protagonists of the cell cycle pathway; CDH1, CDC20 and cyclin E.

Structural and biochemical studies have characterised CCT chaperone-substrate binding[1, 2] though only primary features of the underlying system kinetics/thermodynamics have been revealed[3, 4]. This is of great interest as the current analysis of actin folding/unfolding suggests a ‘thermodynamic chaperone’ requirement; unassisted actin folding may be energetically unfavourable.

Experiment
We hypothesise that CCT plays an ‘active’ role in protein folding, coupling specific substrate-binding interactions to ATP binding/hydrolysis-driven conformational changes. To test this model a system has been developed to examine CCT-actin kinetics at the single molecule level to work towards extracting the thermodynamic parameters of chaperone-bound folding reactions.
Individual CCT complexes have been immobilised via DNA tethering, allowing orientation of molecules ~10nm above modified glass substrates. This method accomplishes the dual fundamental requirements of maintaining protein functionality and allowing sampling on CCT folding timescales (>100s). Single molecule substrate interactions (CCT-actin) can then been probed by TIRF microscopy, allowing analysis of binding and folding kinetics.
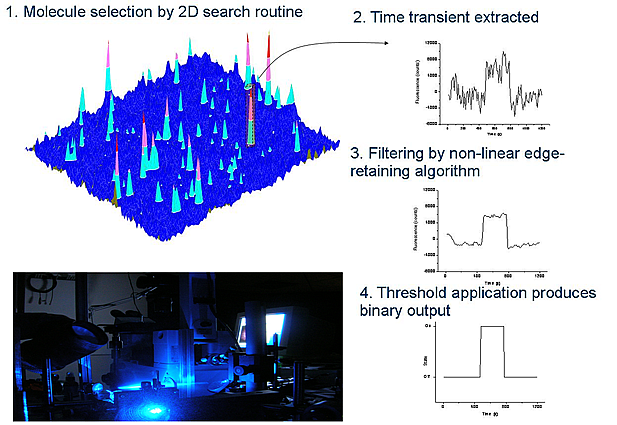
Wide-field microscopy allows simultaneous acquisition of multiple (100s) single molecule interactions. Individual transients can reveal rare events, not observed in ensemble data. Furthermore, analysis of chaperone-substrate lifetime histograms constructed from single interactions allows assess to distribution characteristics, so that details of the reaction energy landscape can be revealed.

Notes:
The chaperones are a class of proteins that assist in the correct folding of inactive, nascent polypeptides to form active proteins. This may require a range of effects including accelerating folding rates, inhibiting off-pathway reactions and preventing aggregation. In all cases the substrate-chaperone interaction is transient and the chaperone does not form part of the final substrate assembly.

TIRF (Total Internal Reflection Fluorescence) microscopy; the excitation laser beam is aligned so as to be incident at the interface between the cover-glass surface and the sample solution at an angle greater than the critical angle. Total internal reflection occurs and a nearfield, standing wave is created, know as the evanescent wave. This penetrates into the sample solution with amplitude that decays exponentially with distance from the interface. Sample excitation by the evanescent wave is thus surface specific (to within ~100nM) and background contribution from excitation of the bulk solution is greatly reduced. TIRF can be achieved via a variety of excitation geometries. In our laboratory the microscope is configured for through-objective TIRF. The excitation beam is directed through the back of the microscope objective, as in traditional epi-illumination, but aligned off-axis so the beam is totally internally reflected at the sample interface, creating the evanescent wave.
Apparatus
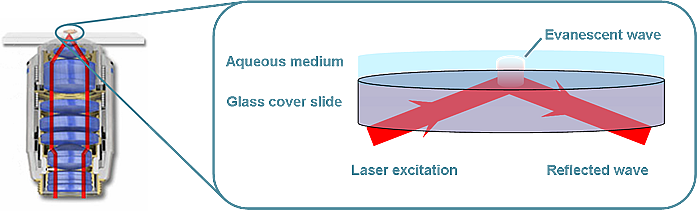
Microscope schematic of through-objective TIRF excitation

References:
1. Llorca, O., et al., Eukaryotic type II chaperonin CCT interacts with actin through specific subunits. Nature, 1999. 402(6762): p. 693-6.
2. McCormack, E.A., M.J. Rohman, and K.R. Willison, Mutational screen identifies critical amino acid residues of beta-actin mediating interaction between its folding intermediates and eukaryotic cytosolic chaperonin CCT. J Struct Biol, 2001. 135(2): p. 185-97.
3. Pappenberger, G., E.A. McCormack, and K.R. Willison, Quantitative actin folding reactions using yeast CCT purified via an internal tag in the CCT3/gamma subunit. J Mol Biol, 2006. 360(2): p. 484-96.
4. Altschuler, G.M., D.R. Klug, and K.R. Willison, Unfolding energetics of G-alpha-actin: a discrete intermediate can be re-folded to the native state by CCT. J Mol Biol, 2005. 353(2): p. 385-96.


|
